|
probeID |
AGICode |
Annotation |
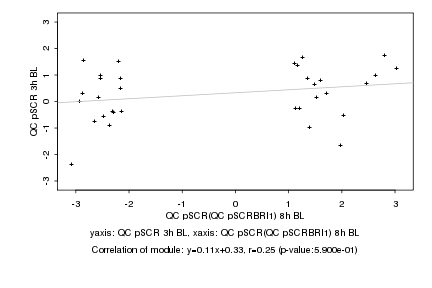
Log2 signal ratio
QC pSCR(QC pSCRBRI1) 8h BL |
Log2 signal ratio
QC pSCR 3h BL |
| 1 |
AT1G60050 |
AT1G60050
|
UMAMIT35, Usually multiple acids move in and out Transporters 35 |
3.023 |
1.250 |
| 2 |
AT2G20570 |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
2.784 |
1.766 |
| 3 |
AT4G37780 |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
2.622 |
0.999 |
| 4 |
AT5G43290 |
AT5G43290
|
ATWRKY49, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 49, WRKY49, WRKY DNA-binding protein 49 |
2.446 |
0.716 |
| 5 |
AT3G48940 |
AT3G48940
|
[Remorin family protein] |
2.013 |
-0.507 |
| 6 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
1.970 |
-1.625 |
| 7 |
AT2G18650 |
AT2G18650
|
MEE16, maternal effect embryo arrest 16 |
1.709 |
0.332 |
| 8 |
AT4G39675 |
AT4G39675
|
unknown |
1.597 |
0.796 |
| 9 |
AT3G62040 |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
1.520 |
0.181 |
| 10 |
AT3G29370 |
AT3G29370
|
P1R3, P1R3 |
1.476 |
0.657 |
| 11 |
AT3G12220 |
AT3G12220
|
scpl16, serine carboxypeptidase-like 16 |
1.387 |
-0.961 |
| 12 |
AT5G24105 |
AT5G24105
|
AGP41, arabinogalactan protein 41 |
1.339 |
0.874 |
| 13 |
AT2G44198 |
AT2G44198
|
unknown |
1.246 |
1.687 |
| 14 |
AT2G22122 |
AT2G22122
|
unknown |
1.204 |
-0.244 |
| 15 |
AT2G22460 |
AT2G22460
|
unknown |
1.157 |
1.361 |
| 16 |
AT5G19890 |
AT5G19890
|
[Peroxidase superfamily protein] |
1.123 |
-0.244 |
| 17 |
AT3G20380 |
AT3G20380
|
[TRAF-like family protein] |
1.101 |
1.458 |
| 18 |
AT3G26120 |
AT3G26120
|
terminal EAR1-like 1 |
-3.095 |
-2.377 |
| 19 |
AT5G53190 |
AT5G53190
|
AtSWEET3, SWEET3 |
-2.940 |
0.006 |
| 20 |
AT5G24110 |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
-2.883 |
0.310 |
| 21 |
AT2G03210 |
AT2G03210
|
ATFUT2, FUT2, fucosyltransferase 2 |
-2.873 |
1.562 |
| 22 |
AT3G46613 |
AT3G46613
|
DVL15, DEVIL 15, RTFL4, ROTUNDIFOLIA like 4 |
-2.655 |
-0.732 |
| 23 |
AT5G66816 |
AT5G66816
|
unknown |
-2.581 |
0.162 |
| 24 |
AT3G15370 |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-2.553 |
0.999 |
| 25 |
AT3G46680 |
AT3G46680
|
[UDP-Glycosyltransferase superfamily protein] |
-2.544 |
0.899 |
| 26 |
AT4G39404 |
AT4G39404
|
other RNA |
-2.488 |
-0.532 |
| 27 |
AT1G17345 |
AT1G17345
|
SAUR77, SMALL AUXIN UPREGULATED RNA 77 |
-2.373 |
-0.905 |
| 28 |
AT5G64990 |
AT5G64990
|
AtRABH1a, RAB GTPase homolog H1A, RABH1a, RAB GTPase homolog H1A |
-2.316 |
-0.343 |
| 29 |
AT3G02150 |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
-2.293 |
-0.389 |
| 30 |
AT1G08090 |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
-2.213 |
1.519 |
| 31 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-2.176 |
0.524 |
| 32 |
AT1G69920 |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
-2.162 |
0.871 |
| 33 |
AT1G23090 |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-2.152 |
-0.346 |