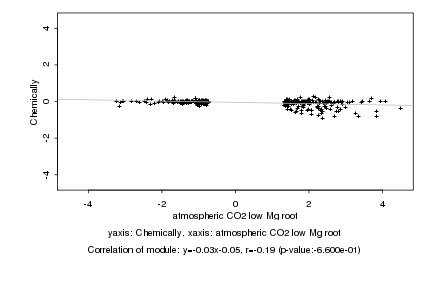
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atmospheric CO2 low Mg root |
Log2 signal ratio
Chemically |
| 1 |
AT4G25220 |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
4.478 |
-0.380 |
| 2 |
AT2G04050 |
AT2G04050
|
[MATE efflux family protein] |
4.060 |
0.036 |
| 3 |
AT3G54530 |
AT3G54530
|
unknown |
3.954 |
no data |
| 4 |
AT3G46240 |
AT3G46240
|
unknown |
3.941 |
0.038 |
| 5 |
AT5G24313 |
AT5G24313
|
unknown |
3.823 |
-0.529 |
| 6 |
AT1G34510 |
AT1G34510
|
[Peroxidase superfamily protein] |
3.816 |
-0.817 |
| 7 |
AT1G02570 |
AT1G02570
|
unknown |
3.731 |
no data |
| 8 |
AT1G05680 |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
3.684 |
0.167 |
| 9 |
AT1G73510 |
AT1G73510
|
unknown |
3.635 |
0.050 |
| 10 |
AT5G43450 |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
3.438 |
0.027 |
| 11 |
AT4G02170 |
AT4G02170
|
unknown |
3.423 |
-0.015 |
| 12 |
AT3G04700 |
AT3G04700
|
unknown |
3.365 |
no data |
| 13 |
AT4G28850 |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
3.335 |
-0.766 |
| 14 |
AT1G54970 |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
3.241 |
-0.626 |
| 15 |
AT5G57200 |
AT5G57200
|
[ENTH/ANTH/VHS superfamily protein] |
3.175 |
0.005 |
| 16 |
AT2G18680 |
AT2G18680
|
unknown |
3.152 |
no data |
| 17 |
AT5G56795 |
AT5G56795
|
MT1B, metallothionein 1B |
3.105 |
no data |
| 18 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
3.092 |
-0.035 |
| 19 |
AT3G20300 |
AT3G20300
|
unknown |
3.032 |
-0.037 |
| 20 |
AT3G07490 |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
2.975 |
-0.285 |
| 21 |
AT2G04515 |
AT2G04515
|
unknown |
2.948 |
no data |
| 22 |
AT4G13390 |
AT4G13390
|
EXT12, extensin 12 |
2.911 |
-0.132 |
| 23 |
AT1G51870 |
AT1G51870
|
[protein kinase family protein] |
2.906 |
no data |
| 24 |
AT1G76210 |
AT1G76210
|
unknown |
2.892 |
0.037 |
| 25 |
AT2G20625 |
AT2G20625
|
unknown |
2.876 |
-0.019 |
| 26 |
AT2G14760 |
AT2G14760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
2.874 |
no data |
| 27 |
AT1G19210 |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
2.849 |
0.021 |
| 28 |
AT4G02270 |
AT4G02270
|
RHS13, root hair specific 13 |
2.834 |
-0.428 |
| 29 |
AT1G50750 |
AT1G50750
|
[Plant mobile domain protein family] |
2.793 |
0.020 |
| 30 |
AT5G43590 |
AT5G43590
|
[Acyl transferase/acyl hydrolase/lysophospholipase superfamily protein] |
2.787 |
-0.053 |
| 31 |
AT3G15420 |
AT3G15420
|
[Transcription factor TFIIIC, tau55-related protein] |
2.786 |
-0.002 |
| 32 |
AT3G62680 |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
2.782 |
-0.513 |
| 33 |
AT3G25655 |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
2.773 |
0.019 |
| 34 |
AT2G05441 |
AT2G05441
|
|
2.763 |
no data |
| 35 |
AT5G06640 |
AT5G06640
|
EXT10, extensin 10 |
2.750 |
-0.284 |
| 36 |
AT3G10710 |
AT3G10710
|
RHS12, root hair specific 12 |
2.747 |
-0.534 |
| 37 |
AT5G55150 |
AT5G55150
|
unknown |
2.709 |
no data |
| 38 |
AT1G76640 |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
2.689 |
-0.045 |
| 39 |
AT1G30800 |
AT1G30800
|
[Fasciclin-like arabinogalactan family protein] |
2.689 |
-0.004 |
| 40 |
AT2G20520 |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
2.682 |
-0.780 |
| 41 |
AT1G18310 |
AT1G18310
|
[glycosyl hydrolase family 81 protein] |
2.667 |
-0.022 |
| 42 |
AT2G33460 |
AT2G33460
|
RIC1, ROP-interactive CRIB motif-containing protein 1 |
2.666 |
-0.066 |
| 43 |
AT2G29620 |
AT2G29620
|
unknown |
2.651 |
-0.098 |
| 44 |
AT5G61550 |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
2.611 |
-0.164 |
| 45 |
AT1G12610 |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
2.608 |
-0.009 |
| 46 |
AT4G14860 |
AT4G14860
|
AtOFP11, ovate family protein 11, OFP11, ovate family protein 11 |
2.573 |
no data |
| 47 |
AT2G31860 |
AT2G31860
|
[pseudogene] |
2.567 |
-0.000 |
| 48 |
AT2G25240 |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
2.562 |
-0.406 |
| 49 |
AT1G33930 |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
2.546 |
0.034 |
| 50 |
AT4G37370 |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
2.538 |
0.228 |
| 51 |
AT5G06990 |
AT5G06990
|
unknown |
2.510 |
no data |
| 52 |
AT1G63030 |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
2.504 |
0.019 |
| 53 |
AT2G43510 |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
2.500 |
0.038 |
| 54 |
AT2G41730 |
AT2G41730
|
unknown |
2.468 |
-0.078 |
| 55 |
AT2G38823 |
AT2G38823
|
unknown |
2.465 |
no data |
| 56 |
AT3G46370 |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
2.459 |
-0.060 |
| 57 |
AT1G74930 |
AT1G74930
|
ORA47 |
2.455 |
-0.020 |
| 58 |
AT1G70460 |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
2.450 |
-0.331 |
| 59 |
AT5G51860 |
AT5G51860
|
AGL72, AGAMOUS-like 72 |
2.442 |
0.074 |
| 60 |
AT1G34540 |
AT1G34540
|
CYP94D1, cytochrome P450, family 94, subfamily D, polypeptide 1 |
2.423 |
-0.220 |
| 61 |
AT3G56790 |
AT3G56790
|
[RNA splicing factor-related] |
2.417 |
no data |
| 62 |
AT1G09080 |
AT1G09080
|
BIP3, binding protein 3 |
2.405 |
-0.077 |
| 63 |
AT5G06630 |
AT5G06630
|
[proline-rich extensin-like family protein] |
2.395 |
-0.242 |
| 64 |
AT3G60280 |
AT3G60280
|
UCC3, uclacyanin 3 |
2.385 |
-0.098 |
| 65 |
AT4G33790 |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
2.369 |
-0.055 |
| 66 |
AT3G49960 |
AT3G49960
|
[Peroxidase superfamily protein] |
2.358 |
-0.523 |
| 67 |
AT5G49080 |
AT5G49080
|
EXT11, extensin 11 |
2.347 |
no data |
| 68 |
AT5G35190 |
AT5G35190
|
EXT13, extensin 13 |
2.346 |
-0.909 |
| 69 |
AT3G54520 |
AT3G54520
|
unknown |
2.344 |
0.023 |
| 70 |
AT4G14280 |
AT4G14280
|
[ARM repeat superfamily protein] |
2.344 |
no data |
| 71 |
AT4G25820 |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
2.335 |
-0.421 |
| 72 |
AT5G04960 |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
2.327 |
-0.621 |
| 73 |
AT4G30320 |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
2.313 |
-0.483 |
| 74 |
AT4G21940 |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
2.309 |
0.002 |
| 75 |
AT2G42850 |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
2.284 |
-0.091 |
| 76 |
AT4G15310 |
AT4G15310
|
CYP702A3, cytochrome P450, family 702, subfamily A, polypeptide 3 |
2.282 |
0.069 |
| 77 |
AT3G52720 |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
2.255 |
0.152 |
| 78 |
AT5G22410 |
AT5G22410
|
RHS18, root hair specific 18 |
2.255 |
-0.738 |
| 79 |
AT4G08410 |
AT4G08410
|
[Proline-rich extensin-like family protein] |
2.247 |
-0.240 |
| 80 |
AT1G14960 |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
2.242 |
-0.356 |
| 81 |
AT1G69240 |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
2.228 |
-0.269 |
| 82 |
AT5G19800 |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
2.225 |
-0.279 |
| 83 |
AT1G32928 |
AT1G32928
|
unknown |
2.186 |
no data |
| 84 |
AT4G22940 |
AT4G22940
|
[Protein kinase superfamily protein] |
2.186 |
-0.006 |
| 85 |
AT1G73120 |
AT1G73120
|
unknown |
2.169 |
0.263 |
| 86 |
AT1G78230 |
AT1G78230
|
[Outer arm dynein light chain 1 protein] |
2.162 |
no data |
| 87 |
AT2G34000 |
AT2G34000
|
[RING/U-box superfamily protein] |
2.157 |
no data |
| 88 |
AT5G58680 |
AT5G58680
|
[ARM repeat superfamily protein] |
2.156 |
-0.007 |
| 89 |
AT2G41380 |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.120 |
0.303 |
| 90 |
AT1G61950 |
AT1G61950
|
CPK19, calcium-dependent protein kinase 19 |
2.109 |
-0.000 |
| 91 |
AT1G06120 |
AT1G06120
|
[Fatty acid desaturase family protein] |
2.086 |
no data |
| 92 |
AT1G07160 |
AT1G07160
|
[Protein phosphatase 2C family protein] |
2.086 |
0.083 |
| 93 |
AT2G37530 |
AT2G37530
|
unknown |
2.080 |
no data |
| 94 |
AT4G26010 |
AT4G26010
|
[Peroxidase superfamily protein] |
2.051 |
-0.465 |
| 95 |
AT4G34580 |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
2.049 |
-0.484 |
| 96 |
AT4G40090 |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
2.048 |
-0.685 |
| 97 |
AT1G05240 |
AT1G05240
|
[Peroxidase superfamily protein] |
2.039 |
no data |
| 98 |
AT4G05049 |
AT4G05049
|
|
2.028 |
no data |
| 99 |
AT1G64930 |
AT1G64930
|
CYP89A7, cytochrome P450, family 87, subfamily A, polypeptide 7 |
2.027 |
-0.039 |
| 100 |
AT1G12150 |
AT1G12150
|
unknown |
2.018 |
-0.088 |
| 101 |
AT4G18830 |
AT4G18830
|
ATOFP5, OFP5, ovate family protein 5 |
2.005 |
no data |
| 102 |
AT3G09925 |
AT3G09925
|
[Pollen Ole e 1 allergen and extensin family protein] |
2.002 |
no data |
| 103 |
AT4G07960 |
AT4G07960
|
ATCSLC12, Cellulose-synthase-like C12, CSLC12, CELLULOSE-SYNTHASE LIKE C12, CSLC12, Cellulose-synthase-like C12 |
1.993 |
-0.145 |
| 104 |
AT1G11170 |
AT1G11170
|
unknown |
1.993 |
0.112 |
| 105 |
AT3G48940 |
AT3G48940
|
[Remorin family protein] |
1.991 |
-0.044 |
| 106 |
AT4G08400 |
AT4G08400
|
[Proline-rich extensin-like family protein] |
1.989 |
no data |
| 107 |
AT4G38420 |
AT4G38420
|
sks9, SKU5 similar 9 |
1.986 |
-0.036 |
| 108 |
AT4G25490 |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
1.985 |
0.022 |
| 109 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
1.980 |
0.123 |
| 110 |
AT5G67400 |
AT5G67400
|
RHS19, root hair specific 19 |
1.976 |
-0.418 |
| 111 |
AT5G51270 |
AT5G51270
|
[U-box domain-containing protein kinase family protein] |
1.975 |
-0.089 |
| 112 |
AT5G43230 |
AT5G43230
|
unknown |
1.971 |
-0.147 |
| 113 |
AT1G05250 |
AT1G05250
|
AtPRX1, PRX2, peroxidase 2 |
1.962 |
no data |
| 114 |
AT5G19560 |
AT5G19560
|
ATROPGEF10, ARABIDOPSIS THALIANA ROP UANINE NUCLEOTIDE EXCHANGE FACTOR 10, GEF10, guanine exchange factor 10, ROPGEF10, ROP (rho of plants) guanine nucleotide exchange factor 10 |
1.957 |
-0.003 |
| 115 |
AT3G11050 |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
1.945 |
-0.058 |
| 116 |
AT5G26320 |
AT5G26320
|
[TRAF-like family protein] |
1.942 |
no data |
| 117 |
AT1G48930 |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
1.941 |
-0.456 |
| 118 |
AT1G10550 |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
1.937 |
0.081 |
| 119 |
AT1G34050 |
AT1G34050
|
[Ankyrin repeat family protein] |
1.917 |
no data |
| 120 |
AT4G01360 |
AT4G01360
|
BPS3, BYPASS 3 |
1.914 |
no data |
| 121 |
AT5G44130 |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
1.907 |
-0.003 |
| 122 |
AT5G46370 |
AT5G46370
|
ATKCO2, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 2, ATTPK2, TANDEM PORE K+ CHANNEL 2, KCO2, Ca2+ activated outward rectifying K+ channel 2 |
1.902 |
-0.005 |
| 123 |
AT3G25950 |
AT3G25950
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
1.900 |
0.013 |
| 124 |
AT3G56010 |
AT3G56010
|
unknown |
1.897 |
-0.014 |
| 125 |
AT4G19760 |
AT4G19760
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
1.877 |
no data |
| 126 |
AT1G27110 |
AT1G27110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
1.869 |
no data |
| 127 |
AT4G33880 |
AT4G33880
|
RSL2, ROOT HAIR DEFECTIVE 6-LIKE 2 |
1.861 |
no data |
| 128 |
AT2G40240 |
AT2G40240
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
1.855 |
0.029 |
| 129 |
AT2G18670 |
AT2G18670
|
[RING/U-box superfamily protein] |
1.855 |
-0.029 |
| 130 |
AT1G34060 |
AT1G34060
|
[Pyridoxal phosphate (PLP)-dependent transferases superfamily protein] |
1.843 |
no data |
| 131 |
AT1G79860 |
AT1G79860
|
ATROPGEF12, MEE64, MATERNAL EFFECT EMBRYO ARREST 64, ROPGEF12, ROP (rho of plants) guanine nucleotide exchange factor 12 |
1.841 |
-0.114 |
| 132 |
AT4G20160 |
AT4G20160
|
[LOCATED IN: chloroplast] |
1.840 |
-0.030 |
| 133 |
AT2G29010 |
AT2G29010
|
[pseudogene] |
1.839 |
no data |
| 134 |
AT1G12040 |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
1.835 |
-0.315 |
| 135 |
AT1G65570 |
AT1G65570
|
[Pectin lyase-like superfamily protein] |
1.828 |
-0.163 |
| 136 |
AT3G07720 |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
1.827 |
0.045 |
| 137 |
AT3G01830 |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
1.820 |
0.040 |
| 138 |
AT2G34180 |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
1.820 |
-0.219 |
| 139 |
AT2G18193 |
AT2G18193
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.796 |
no data |
| 140 |
AT5G57625 |
AT5G57625
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.785 |
no data |
| 141 |
AT5G38970 |
AT5G38970
|
ATBR6OX, BR6OX, BRASSINOSTEROID-6-OXIDASE, BR6OX1, brassinosteroid-6-oxidase 1, CYP85A1 |
1.784 |
0.021 |
| 142 |
AT1G36622 |
AT1G36622
|
unknown |
1.783 |
no data |
| 143 |
AT1G19900 |
AT1G19900
|
[glyoxal oxidase-related protein] |
1.778 |
-0.046 |
| 144 |
AT4G00680 |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
1.776 |
-0.650 |
| 145 |
AT4G33730 |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.770 |
-0.439 |
| 146 |
AT1G78340 |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
1.758 |
-0.056 |
| 147 |
AT1G02610 |
AT1G02610
|
[RING/FYVE/PHD zinc finger superfamily protein] |
1.756 |
no data |
| 148 |
AT3G14850 |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
1.743 |
0.250 |
| 149 |
AT5G13210 |
AT5G13210
|
[Uncharacterised conserved protein UCP015417, vWA] |
1.734 |
no data |
| 150 |
AT3G06750 |
AT3G06750
|
[hydroxyproline-rich glycoprotein family protein] |
1.734 |
-0.036 |
| 151 |
AT5G36260 |
AT5G36260
|
[Eukaryotic aspartyl protease family protein] |
1.708 |
-0.045 |
| 152 |
AT5G61350 |
AT5G61350
|
[Protein kinase superfamily protein] |
1.705 |
-0.236 |
| 153 |
AT4G13420 |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
1.697 |
0.028 |
| 154 |
AT5G11140 |
AT5G11140
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
1.682 |
no data |
| 155 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
1.677 |
-0.042 |
| 156 |
AT5G56544 |
AT5G56544
|
|
1.672 |
no data |
| 157 |
AT2G17590 |
AT2G17590
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.672 |
-0.001 |
| 158 |
AT1G73530 |
AT1G73530
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.671 |
-0.007 |
| 159 |
AT5G55050 |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.670 |
0.129 |
| 160 |
AT1G30870 |
AT1G30870
|
[Peroxidase superfamily protein] |
1.670 |
-0.356 |
| 161 |
AT4G29780 |
AT4G29780
|
unknown |
1.669 |
0.025 |
| 162 |
AT2G03760 |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
1.668 |
0.022 |
| 163 |
AT4G39070 |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
1.648 |
-0.061 |
| 164 |
AT5G41160 |
AT5G41160
|
ATPUP12, ARABIDOPSIS THALIANA PURINE PERMEASE 12, PUP12, purine permease 12 |
1.645 |
no data |
| 165 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
1.645 |
0.010 |
| 166 |
AT5G38700 |
AT5G38700
|
unknown |
1.644 |
-0.089 |
| 167 |
AT1G80800 |
AT1G80800
|
[pseudogene] |
1.644 |
0.031 |
| 168 |
AT2G03720 |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
1.643 |
-0.522 |
| 169 |
AT3G58810 |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
1.632 |
-0.032 |
| 170 |
AT4G25790 |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.631 |
-0.547 |
| 171 |
AT5G46870 |
AT5G46870
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.610 |
no data |
| 172 |
AT2G28270 |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.608 |
0.061 |
| 173 |
AT2G41640 |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
1.606 |
-0.021 |
| 174 |
AT2G43590 |
AT2G43590
|
[Chitinase family protein] |
1.602 |
0.084 |
| 175 |
AT4G22080 |
AT4G22080
|
RHS14, root hair specific 14 |
1.579 |
no data |
| 176 |
AT1G52660 |
AT1G52660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.572 |
-0.134 |
| 177 |
AT3G08040 |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
1.569 |
-0.000 |
| 178 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
1.563 |
0.013 |
| 179 |
AT1G60190 |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
1.562 |
-0.018 |
| 180 |
AT1G74450 |
AT1G74450
|
unknown |
1.545 |
-0.001 |
| 181 |
AT1G45140 |
AT1G45140
|
[non-LTR retrotransposon family (LINE), has a 1.4e-37 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
1.537 |
no data |
| 182 |
AT2G30020 |
AT2G30020
|
[Protein phosphatase 2C family protein] |
1.537 |
no data |
| 183 |
AT2G35890 |
AT2G35890
|
CPK25, calcium-dependent protein kinase 25 |
1.537 |
-0.010 |
| 184 |
AT5G26038 |
AT5G26038
|
MIR860A, microRNA860A |
1.531 |
no data |
| 185 |
AT3G22830 |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
1.530 |
-0.047 |
| 186 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
1.523 |
0.022 |
| 187 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
1.517 |
-0.125 |
| 188 |
AT2G44110 |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
1.511 |
-0.467 |
| 189 |
AT1G18300 |
AT1G18300
|
atnudt4, nudix hydrolase homolog 4, NUDT4, nudix hydrolase homolog 4 |
1.505 |
no data |
| 190 |
AT5G55110 |
AT5G55110
|
[Stigma-specific Stig1 family protein] |
1.504 |
no data |
| 191 |
AT5G57560 |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
1.504 |
-0.031 |
| 192 |
AT5G04950 |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
1.501 |
-0.144 |
| 193 |
AT1G12560 |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
1.474 |
-0.420 |
| 194 |
AT2G46400 |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
1.465 |
0.054 |
| 195 |
AT5G15970 |
AT5G15970
|
AtCor6.6, COR6.6, COLD-RESPONSIVE 6.6, KIN2 |
1.465 |
no data |
| 196 |
AT3G26490 |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
1.465 |
0.006 |
| 197 |
AT1G56660 |
AT1G56660
|
unknown |
1.464 |
0.001 |
| 198 |
AT1G65310 |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
1.463 |
0.130 |
| 199 |
AT1G63930 |
AT1G63930
|
ROH1, from the Czech 'roh' meaning 'corner' |
1.455 |
-0.059 |
| 200 |
AT5G42590 |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
1.440 |
-0.032 |
| 201 |
AT2G21010 |
AT2G21010
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
1.436 |
no data |
| 202 |
AT5G20850 |
AT5G20850
|
ATRAD51, RAD51 |
1.435 |
0.033 |
| 203 |
AT4G19690 |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
1.427 |
-0.137 |
| 204 |
AT2G33710 |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
1.423 |
0.076 |
| 205 |
AT5G19790 |
AT5G19790
|
RAP2.11, related to AP2 11 |
1.420 |
-0.269 |
| 206 |
AT4G31940 |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
1.419 |
0.032 |
| 207 |
AT3G45070 |
AT3G45070
|
AtSULT202B1, SULT202B1, sulfotransferase 202B1 |
1.419 |
no data |
| 208 |
AT2G20080 |
AT2G20080
|
TIE2, TCP Interactor containing EAR motif protein 2 |
1.411 |
no data |
| 209 |
AT4G12520 |
AT4G12520
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.406 |
no data |
| 210 |
AT1G62980 |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
1.404 |
-0.402 |
| 211 |
AT2G41970 |
AT2G41970
|
[Protein kinase superfamily protein] |
1.404 |
-0.250 |
| 212 |
AT1G16225 |
AT1G16225
|
[Target SNARE coiled-coil domain protein] |
1.403 |
no data |
| 213 |
AT1G76800 |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
1.401 |
-0.106 |
| 214 |
AT1G70080 |
AT1G70080
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
1.399 |
-0.023 |
| 215 |
AT1G08430 |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
1.392 |
0.162 |
| 216 |
AT1G18740 |
AT1G18740
|
unknown |
1.389 |
-0.028 |
| 217 |
AT5G11180 |
AT5G11180
|
ATGLR2.6, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.6, GLR2.6, glutamate receptor 2.6 |
1.387 |
-0.012 |
| 218 |
AT4G22666 |
AT4G22666
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.383 |
no data |
| 219 |
AT3G25250 |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
1.383 |
-0.001 |
| 220 |
AT2G17442 |
AT2G17442
|
unknown |
1.377 |
no data |
| 221 |
AT1G14185 |
AT1G14185
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
1.373 |
no data |
| 222 |
AT2G32090 |
AT2G32090
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
1.372 |
-0.020 |
| 223 |
AT2G23910 |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.367 |
0.151 |
| 224 |
AT1G76220 |
AT1G76220
|
unknown |
1.359 |
0.046 |
| 225 |
AT5G15180 |
AT5G15180
|
[Peroxidase superfamily protein] |
1.358 |
-0.228 |
| 226 |
AT1G19025 |
AT1G19025
|
[DNA repair metallo-beta-lactamase family protein] |
1.358 |
-0.062 |
| 227 |
AT2G20670 |
AT2G20670
|
unknown |
1.356 |
-0.040 |
| 228 |
AT1G51830 |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
1.354 |
-0.157 |
| 229 |
AT5G11070 |
AT5G11070
|
unknown |
1.352 |
-0.071 |
| 230 |
AT5G26300 |
AT5G26300
|
[TRAF-like family protein] |
1.350 |
no data |
| 231 |
AT4G08620 |
AT4G08620
|
SULTR1;1, sulphate transporter 1;1 |
1.349 |
-0.149 |
| 232 |
AT1G73540 |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
1.347 |
-0.031 |
| 233 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
1.345 |
-0.008 |
| 234 |
AT2G21880 |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
1.345 |
-0.190 |
| 235 |
AT5G36180 |
AT5G36180
|
scpl1, serine carboxypeptidase-like 1 |
1.345 |
no data |
| 236 |
AT1G60010 |
AT1G60010
|
unknown |
1.332 |
0.043 |
| 237 |
AT1G13420 |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
1.331 |
-0.186 |
| 238 |
AT5G41800 |
AT5G41800
|
[Transmembrane amino acid transporter family protein] |
1.322 |
-0.010 |
| 239 |
AT1G09170 |
AT1G09170
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
1.321 |
-0.169 |
| 240 |
AT4G25190 |
AT4G25190
|
QWRF7, QWRF domain containing 7 |
1.318 |
no data |
| 241 |
AT5G38930 |
AT5G38930
|
[RmlC-like cupins superfamily protein] |
1.312 |
no data |
| 242 |
AT2G32487 |
AT2G32487
|
unknown |
1.310 |
no data |
| 243 |
AT1G47395 |
AT1G47395
|
unknown |
-3.993 |
no data |
| 244 |
AT1G29100 |
AT1G29100
|
[Heavy metal transport/detoxification superfamily protein ] |
-3.332 |
no data |
| 245 |
AT5G53380 |
AT5G53380
|
[O-acyltransferase (WSD1-like) family protein] |
-3.303 |
no data |
| 246 |
AT5G38430 |
AT5G38430
|
RBCS1B, Rubisco small subunit 1B |
-3.285 |
no data |
| 247 |
AT1G08090 |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
-3.239 |
0.008 |
| 248 |
AT1G07400 |
AT1G07400
|
[HSP20-like chaperones superfamily protein] |
-3.186 |
no data |
| 249 |
AT1G47400 |
AT1G47400
|
unknown |
-3.160 |
-0.243 |
| 250 |
AT2G17850 |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-3.135 |
-0.045 |
| 251 |
AT2G44000 |
AT2G44000
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-3.085 |
0.052 |
| 252 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
-3.063 |
no data |
| 253 |
AT2G13810 |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
-3.053 |
0.015 |
| 254 |
AT4G22214 |
AT4G22214
|
[Defensin-like (DEFL) family protein] |
-3.003 |
no data |
| 255 |
AT5G39610 |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-2.827 |
0.017 |
| 256 |
AT1G29910 |
AT1G29910
|
AB180, CAB3, chlorophyll A/B binding protein 3, LHCB1.2, LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN 1.2 |
-2.800 |
no data |
| 257 |
AT2G30760 |
AT2G30760
|
unknown |
-2.705 |
0.004 |
| 258 |
AT3G50170 |
AT3G50170
|
unknown |
-2.632 |
-0.030 |
| 259 |
AT1G36305 |
AT1G36305
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-2.555 |
no data |
| 260 |
AT5G35870 |
AT5G35870
|
unknown |
-2.474 |
0.048 |
| 261 |
AT1G68010 |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
-2.443 |
-0.015 |
| 262 |
AT3G60420 |
AT3G60420
|
[Phosphoglycerate mutase family protein] |
-2.409 |
0.025 |
| 263 |
AT3G51220 |
AT3G51220
|
unknown |
-2.389 |
no data |
| 264 |
AT3G52480 |
AT3G52480
|
unknown |
-2.333 |
-0.114 |
| 265 |
AT5G18362 |
AT5G18362
|
[Cytokine-induced anti-apoptosis inhibitor 1, Fe-S biogenesis] |
-2.318 |
no data |
| 266 |
AT5G13320 |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-2.287 |
0.128 |
| 267 |
AT5G39180 |
AT5G39180
|
[RmlC-like cupins superfamily protein] |
-2.255 |
no data |
| 268 |
AT4G16230 |
AT4G16230
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-2.246 |
no data |
| 269 |
AT1G01680 |
AT1G01680
|
ATPUB54, PLANT U-BOX 54, PUB54, plant U-box 54 |
-2.225 |
no data |
| 270 |
AT3G46230 |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-2.221 |
-0.093 |
| 271 |
AT5G28650 |
AT5G28650
|
ATWRKY74, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 74, WRKY74, WRKY DNA-binding protein 74 |
-2.116 |
-0.021 |
| 272 |
AT1G66970 |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-2.079 |
0.028 |
| 273 |
AT4G01390 |
AT4G01390
|
[TRAF-like family protein] |
-1.992 |
0.030 |
| 274 |
AT1G06680 |
AT1G06680
|
OE23, OXYGEN EVOLVING COMPLEX SUBUNIT 23 KDA, OEE2, OXYGEN-EVOLVING ENHANCER PROTEIN 2, PSBP-1, photosystem II subunit P-1, PSII-P, PHOTOSYSTEM II SUBUNIT P |
-1.957 |
0.001 |
| 275 |
AT5G41690 |
AT5G41690
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-1.942 |
no data |
| 276 |
AT3G50770 |
AT3G50770
|
CML41, calmodulin-like 41 |
-1.922 |
0.143 |
| 277 |
AT2G25605 |
AT2G25605
|
unknown |
-1.918 |
no data |
| 278 |
AT2G27400 |
AT2G27400
|
TAS1A, trans-acting siRNA1A |
-1.894 |
no data |
| 279 |
AT5G62330 |
AT5G62330
|
unknown |
-1.870 |
0.059 |
| 280 |
AT1G67000 |
AT1G67000
|
[Protein kinase superfamily protein] |
-1.848 |
0.050 |
| 281 |
AT1G09310 |
AT1G09310
|
unknown |
-1.824 |
-0.009 |
| 282 |
AT3G28510 |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.794 |
0.007 |
| 283 |
AT1G26200 |
AT1G26200
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-1.767 |
no data |
| 284 |
AT2G26150 |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-1.738 |
0.020 |
| 285 |
AT3G55090 |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
-1.738 |
-0.011 |
| 286 |
AT5G52860 |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
-1.736 |
0.051 |
| 287 |
AT4G10120 |
AT4G10120
|
ATSPS4F |
-1.722 |
0.100 |
| 288 |
AT2G32690 |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-1.707 |
-0.105 |
| 289 |
AT4G22790 |
AT4G22790
|
[MATE efflux family protein] |
-1.695 |
0.048 |
| 290 |
AT1G18970 |
AT1G18970
|
GLP4, germin-like protein 4 |
-1.689 |
-0.037 |
| 291 |
AT5G41280 |
AT5G41280
|
[Receptor-like protein kinase-related family protein] |
-1.679 |
-0.043 |
| 292 |
AT2G32860 |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-1.673 |
0.106 |
| 293 |
AT1G06080 |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-1.667 |
0.244 |
| 294 |
AT2G36305 |
AT2G36305
|
ATFACE-2, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN-CONVERTING ENZYME-2, ATFACE2, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN-CONVERTING ENZYME 2, FACE2, farnesylated protein-converting enzyme 2, RCE1, RAS-CONVERTING ENZYME 1 |
-1.638 |
no data |
| 295 |
AT3G50400 |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.633 |
0.003 |
| 296 |
AT4G16220 |
AT4G16220
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.632 |
no data |
| 297 |
AT5G66670 |
AT5G66670
|
unknown |
-1.588 |
-0.006 |
| 298 |
AT2G34315 |
AT2G34315
|
[Avirulence induced gene (AIG1) family protein] |
-1.553 |
0.015 |
| 299 |
AT3G21890 |
AT3G21890
|
BBX31, B-box domain protein 31 |
-1.542 |
-0.019 |
| 300 |
AT3G59930 |
AT3G59930
|
unknown |
-1.542 |
no data |
| 301 |
AT2G38110 |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-1.518 |
-0.055 |
| 302 |
AT1G47620 |
AT1G47620
|
CYP96A8, cytochrome P450, family 96, subfamily A, polypeptide 8 |
-1.517 |
-0.037 |
| 303 |
AT1G27580 |
AT1G27580
|
unknown |
-1.509 |
no data |
| 304 |
AT2G24190 |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-1.509 |
0.008 |
| 305 |
AT5G15030 |
AT5G15030
|
[Paired amphipathic helix (PAH2) superfamily protein] |
-1.495 |
no data |
| 306 |
AT2G25410 |
AT2G25410
|
[RING/U-box superfamily protein] |
-1.481 |
no data |
| 307 |
AT1G17230 |
AT1G17230
|
[Leucine-rich receptor-like protein kinase family protein] |
-1.471 |
0.024 |
| 308 |
AT5G14830 |
AT5G14830
|
[retrotransposon family] |
-1.466 |
0.009 |
| 309 |
AT5G39150 |
AT5G39150
|
[RmlC-like cupins superfamily protein] |
-1.457 |
no data |
| 310 |
AT1G34670 |
AT1G34670
|
AtMYB93, myb domain protein 93, MYB93, myb domain protein 93 |
-1.447 |
-0.125 |
| 311 |
AT4G16370 |
AT4G16370
|
oligopeptide transporter |
-1.434 |
-0.051 |
| 312 |
AT5G05340 |
AT5G05340
|
PRX52, peroxidase 52 |
-1.432 |
0.049 |
| 313 |
AT5G03860 |
AT5G03860
|
MLS, malate synthase |
-1.432 |
-0.095 |
| 314 |
AT1G68530 |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-1.424 |
-0.033 |
| 315 |
AT5G26690 |
AT5G26690
|
[Heavy metal transport/detoxification superfamily protein ] |
-1.421 |
no data |
| 316 |
AT5G49850 |
AT5G49850
|
[Mannose-binding lectin superfamily protein] |
-1.413 |
no data |
| 317 |
AT1G26570 |
AT1G26570
|
ATUGD1, UDP-GLUCOSE DEHYDROGENASE 1, UGD1, UDP-glucose dehydrogenase 1 |
-1.408 |
-0.017 |
| 318 |
AT3G51600 |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-1.388 |
0.000 |
| 319 |
AT2G03200 |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
-1.386 |
-0.024 |
| 320 |
AT2G35380 |
AT2G35380
|
[Peroxidase superfamily protein] |
-1.385 |
-0.036 |
| 321 |
AT2G18370 |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.383 |
0.029 |
| 322 |
AT1G20070 |
AT1G20070
|
unknown |
-1.365 |
0.025 |
| 323 |
AT2G39510 |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-1.356 |
0.021 |
| 324 |
AT1G30040 |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
-1.347 |
0.056 |
| 325 |
AT1G29465 |
AT1G29465
|
unknown |
-1.346 |
no data |
| 326 |
AT2G43670 |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
-1.341 |
-0.074 |
| 327 |
AT1G01600 |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-1.338 |
0.005 |
| 328 |
AT4G10500 |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.332 |
0.026 |
| 329 |
AT5G16990 |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
-1.326 |
-0.014 |
| 330 |
AT1G64170 |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-1.315 |
0.097 |
| 331 |
AT1G65860 |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-1.290 |
-0.066 |
| 332 |
AT1G77700 |
AT1G77700
|
[Pathogenesis-related thaumatin superfamily protein] |
-1.280 |
0.023 |
| 333 |
AT1G53560 |
AT1G53560
|
[Ribosomal protein L18ae family] |
-1.272 |
-0.006 |
| 334 |
AT4G25860 |
AT4G25860
|
ORP4A, OSBP(oxysterol binding protein)-related protein 4A |
-1.272 |
no data |
| 335 |
AT1G23200 |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-1.270 |
-0.014 |
| 336 |
AT2G21900 |
AT2G21900
|
ATWRKY59, WRKY59, WRKY DNA-binding protein 59 |
-1.270 |
no data |
| 337 |
AT2G47670 |
AT2G47670
|
PMEI6, PECTIN METHYLESTERASE INHIBITOR 6 |
-1.269 |
0.053 |
| 338 |
AT1G52620 |
AT1G52620
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.258 |
0.000 |
| 339 |
AT3G54070 |
AT3G54070
|
[Ankyrin repeat family protein] |
-1.257 |
0.043 |
| 340 |
AT1G03270 |
AT1G03270
|
unknown |
-1.252 |
0.019 |
| 341 |
AT1G66520 |
AT1G66520
|
pde194, pigment defective 194 |
-1.242 |
-0.018 |
| 342 |
AT3G58940 |
AT3G58940
|
[F-box/RNI-like superfamily protein] |
-1.215 |
no data |
| 343 |
AT5G58784 |
AT5G58784
|
[Undecaprenyl pyrophosphate synthetase family protein] |
-1.212 |
-0.034 |
| 344 |
AT1G60470 |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
-1.208 |
0.020 |
| 345 |
AT1G19960 |
AT1G19960
|
unknown |
-1.196 |
0.017 |
| 346 |
AT1G73470 |
AT1G73470
|
unknown |
-1.196 |
-0.013 |
| 347 |
AT5G62200 |
AT5G62200
|
[Embryo-specific protein 3, (ATS3)] |
-1.192 |
0.025 |
| 348 |
AT4G37445 |
AT4G37445
|
unknown |
-1.183 |
0.039 |
| 349 |
AT5G56720 |
AT5G56720
|
c-NAD-MDH3, cytosolic-NAD-dependent malate dehydrogenase 3 |
-1.156 |
0.042 |
| 350 |
AT5G48335 |
AT5G48335
|
unknown |
-1.127 |
no data |
| 351 |
AT3G47600 |
AT3G47600
|
ATMYB94, myb domain protein 94, ATMYBCP70, MYB94, myb domain protein 94 |
-1.120 |
-0.030 |
| 352 |
AT3G10070 |
AT3G10070
|
TAF12, TBP-associated factor 12, TAFII58, TATA-ASSOCIATED FACTOR II 58 |
-1.115 |
-0.002 |
| 353 |
AT5G65140 |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-1.105 |
-0.028 |
| 354 |
AT1G64160 |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-1.101 |
0.189 |
| 355 |
AT1G19250 |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-1.098 |
0.172 |
| 356 |
AT4G36190 |
AT4G36190
|
[Serine carboxypeptidase S28 family protein] |
-1.084 |
no data |
| 357 |
AT3G43430 |
AT3G43430
|
[RING/U-box superfamily protein] |
-1.083 |
-0.031 |
| 358 |
AT2G45530 |
AT2G45530
|
[RING/U-box superfamily protein] |
-1.077 |
-0.031 |
| 359 |
AT1G04220 |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-1.075 |
0.078 |
| 360 |
AT5G13580 |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
-1.066 |
-0.047 |
| 361 |
AT2G26910 |
AT2G26910
|
ABCG32, ATP-binding cassette G32, AtABCG32, ATPDR4, PLEIOTROPIC DRUG RESISTANCE 4, PDR4, pleiotropic drug resistance 4, PEC1, PERMEABLE CUTICLE 1 |
-1.065 |
-0.065 |
| 362 |
AT3G50430 |
AT3G50430
|
unknown |
-1.064 |
-0.015 |
| 363 |
AT2G16630 |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
-1.064 |
-0.022 |
| 364 |
AT1G29660 |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.057 |
-0.117 |
| 365 |
AT4G37060 |
AT4G37060
|
AtPLAIVB, phospholipase A IVB, PLA IVB, PLP5, PATATIN-like protein 5 |
-1.056 |
no data |
| 366 |
AT2G22850 |
AT2G22850
|
AtbZIP6, basic leucine-zipper 6, bZIP6, basic leucine-zipper 6 |
-1.054 |
-0.097 |
| 367 |
ATMG01080 |
ATMG01080
|
ATP9, mitochondrial F0-ATPase subunit 9 |
-1.050 |
no data |
| 368 |
AT2G48140 |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
-1.043 |
-0.083 |
| 369 |
AT2G04800 |
AT2G04800
|
unknown |
-1.039 |
-0.172 |
| 370 |
AT3G23805 |
AT3G23805
|
RALFL24, ralf-like 24 |
-1.039 |
-0.066 |
| 371 |
AT1G20340 |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
-1.038 |
-0.013 |
| 372 |
AT5G53830 |
AT5G53830
|
[VQ motif-containing protein] |
-1.034 |
-0.044 |
| 373 |
AT5G09520 |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-1.032 |
-0.117 |
| 374 |
AT5G11640 |
AT5G11640
|
[Thioredoxin superfamily protein] |
-1.027 |
-0.022 |
| 375 |
AT3G27070 |
AT3G27070
|
TOM20-1, translocase outer membrane 20-1 |
-1.026 |
0.042 |
| 376 |
AT4G14390 |
AT4G14390
|
[Ankyrin repeat family protein] |
-1.025 |
0.077 |
| 377 |
AT5G50200 |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
-1.018 |
0.034 |
| 378 |
AT4G30610 |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-1.004 |
-0.017 |
| 379 |
AT2G23540 |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.999 |
-0.048 |
| 380 |
AT1G73410 |
AT1G73410
|
ATMYB54, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 54, MYB54, myb domain protein 54 |
-0.999 |
-0.027 |
| 381 |
AT5G12420 |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.992 |
-0.091 |
| 382 |
AT4G21903 |
AT4G21903
|
[MATE efflux family protein] |
-0.992 |
no data |
| 383 |
AT4G28940 |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.992 |
-0.224 |
| 384 |
AT5G63690 |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.988 |
0.076 |
| 385 |
AT2G20310 |
AT2G20310
|
RIN13, RPM1 interacting protein 13 |
-0.987 |
-0.007 |
| 386 |
AT5G40240 |
AT5G40240
|
UMAMIT40, Usually multiple acids move in and out Transporters 40 |
-0.984 |
no data |
| 387 |
AT4G16060 |
AT4G16060
|
unknown |
-0.983 |
0.038 |
| 388 |
AT2G47560 |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.979 |
0.068 |
| 389 |
AT3G17050 |
AT3G17050
|
[pseudogene] |
-0.976 |
no data |
| 390 |
AT1G13930 |
AT1G13930
|
unknown |
-0.973 |
-0.000 |
| 391 |
AT3G15650 |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.970 |
-0.050 |
| 392 |
AT5G19370 |
AT5G19370
|
[rhodanese-like domain-containing protein / PPIC-type PPIASE domain-containing protein] |
-0.965 |
0.003 |
| 393 |
AT1G73600 |
AT1G73600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.948 |
no data |
| 394 |
AT1G73602 |
AT1G73602
|
CPuORF32, conserved peptide upstream open reading frame 32 |
-0.948 |
no data |
| 395 |
AT4G32860 |
AT4G32860
|
unknown |
-0.946 |
-0.109 |
| 396 |
AT1G78770 |
AT1G78770
|
APC6, anaphase promoting complex 6 |
-0.943 |
0.081 |
| 397 |
AT2G13840 |
AT2G13840
|
[Polymerase/histidinol phosphatase-like] |
-0.936 |
0.025 |
| 398 |
AT1G63180 |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.935 |
-0.019 |
| 399 |
AT2G39890 |
AT2G39890
|
ATPROT1, PROLINE TRANSPORTER 1, PROT1, proline transporter 1 |
-0.934 |
-0.006 |
| 400 |
AT2G36690 |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.931 |
0.036 |
| 401 |
AT3G58180 |
AT3G58180
|
[ARM repeat superfamily protein] |
-0.929 |
0.005 |
| 402 |
AT1G03040 |
AT1G03040
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.926 |
0.036 |
| 403 |
AT5G52710 |
AT5G52710
|
[Copper transport protein family] |
-0.925 |
0.001 |
| 404 |
AT5G06620 |
AT5G06620
|
ATXR4, SDG38, SET domain protein 38 |
-0.919 |
no data |
| 405 |
AT5G20180 |
AT5G20180
|
[Ribosomal protein L36] |
-0.917 |
0.003 |
| 406 |
AT3G18400 |
AT3G18400
|
anac058, NAC domain containing protein 58, NAC058, NAC domain containing protein 58 |
-0.915 |
-0.071 |
| 407 |
AT1G01370 |
AT1G01370
|
CENH3, CENTROMERIC HISTONE H3, HTR12 |
-0.913 |
0.091 |
| 408 |
AT5G41040 |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
-0.909 |
-0.060 |
| 409 |
AT2G25080 |
AT2G25080
|
ATGPX1, GLUTATHIONE PEROXIDASE 1, GPX1, glutathione peroxidase 1 |
-0.908 |
-0.005 |
| 410 |
AT2G41040 |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.906 |
0.013 |
| 411 |
AT2G33830 |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.896 |
-0.020 |
| 412 |
AT3G53510 |
AT3G53510
|
ABCG20, ATP-binding cassette G20 |
-0.896 |
-0.048 |
| 413 |
AT5G50140 |
AT5G50140
|
[Ankyrin repeat family protein] |
-0.895 |
0.009 |
| 414 |
AT2G16800 |
AT2G16800
|
[high-affinity nickel-transport family protein] |
-0.894 |
0.014 |
| 415 |
AT2G24720 |
AT2G24720
|
ATGLR2.2, glutamate receptor 2.2, GLR2.2, glutamate receptor 2.2 |
-0.894 |
no data |
| 416 |
AT3G11430 |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
-0.891 |
-0.144 |
| 417 |
AT3G12210 |
AT3G12210
|
[DNA binding] |
-0.887 |
0.019 |
| 418 |
AT5G24220 |
AT5G24220
|
[Lipase class 3-related protein] |
-0.885 |
no data |
| 419 |
AT5G17610 |
AT5G17610
|
unknown |
-0.882 |
0.012 |
| 420 |
AT1G49360 |
AT1G49360
|
[F-box family protein] |
-0.881 |
no data |
| 421 |
AT5G66320 |
AT5G66320
|
GATA5, GATA transcription factor 5 |
-0.878 |
0.023 |
| 422 |
AT2G42670 |
AT2G42670
|
unknown |
-0.877 |
0.016 |
| 423 |
AT5G18240 |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.877 |
0.058 |
| 424 |
AT4G23730 |
AT4G23730
|
[Galactose mutarotase-like superfamily protein] |
-0.875 |
-0.018 |
| 425 |
AT2G42770 |
AT2G42770
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.875 |
-0.007 |
| 426 |
AT5G18245 |
AT5G18245
|
other RNA |
-0.872 |
no data |
| 427 |
AT1G08160 |
AT1G08160
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.869 |
no data |
| 428 |
AT3G50350 |
AT3G50350
|
unknown |
-0.865 |
0.001 |
| 429 |
AT5G23920 |
AT5G23920
|
unknown |
-0.861 |
-0.046 |
| 430 |
AT3G15690 |
AT3G15690
|
[Single hybrid motif superfamily protein] |
-0.860 |
-0.019 |
| 431 |
AT2G48130 |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.857 |
-0.063 |
| 432 |
AT1G01080 |
AT1G01080
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.856 |
0.011 |
| 433 |
AT5G54960 |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.853 |
0.004 |
| 434 |
AT3G46560 |
AT3G46560
|
emb2474, embryo defective 2474, TIM9 |
-0.851 |
0.025 |
| 435 |
AT2G30530 |
AT2G30530
|
unknown |
-0.851 |
-0.001 |
| 436 |
AT2G38460 |
AT2G38460
|
ATIREG1, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 1, FPN1, FERROPORTIN 1, IREG1, iron regulated 1 |
-0.848 |
-0.051 |
| 437 |
AT2G37700 |
AT2G37700
|
[Fatty acid hydroxylase superfamily] |
-0.848 |
-0.007 |
| 438 |
AT3G16370 |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.846 |
-0.049 |
| 439 |
AT3G14200 |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.845 |
0.020 |
| 440 |
AT4G12710 |
AT4G12710
|
[ARM repeat superfamily protein] |
-0.843 |
no data |
| 441 |
AT1G05880 |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
-0.842 |
0.039 |
| 442 |
AT1G01190 |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.842 |
-0.076 |
| 443 |
AT5G23190 |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.842 |
-0.139 |
| 444 |
AT1G52140 |
AT1G52140
|
unknown |
-0.840 |
0.009 |
| 445 |
AT5G37450 |
AT5G37450
|
[Leucine-rich repeat protein kinase family protein] |
-0.837 |
0.053 |
| 446 |
AT5G54130 |
AT5G54130
|
[Calcium-binding endonuclease/exonuclease/phosphatase family] |
-0.837 |
0.018 |
| 447 |
AT2G40190 |
AT2G40190
|
LEW3, LEAF WILTING 3 |
-0.831 |
no data |
| 448 |
AT4G16920 |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.830 |
-0.116 |
| 449 |
AT5G14760 |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.828 |
-0.045 |
| 450 |
AT5G19530 |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.827 |
0.061 |
| 451 |
AT2G33860 |
AT2G33860
|
ARF3, AUXIN RESPONSE TRANSCRIPTION FACTOR 3, ETT, ETTIN |
-0.827 |
0.025 |
| 452 |
AT1G62280 |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.825 |
0.068 |
| 453 |
AT1G01180 |
AT1G01180
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.824 |
no data |
| 454 |
AT1G67150 |
AT1G67150
|
unknown |
-0.812 |
no data |
| 455 |
AT3G01740 |
AT3G01740
|
[Mitochondrial ribosomal protein L37] |
-0.808 |
0.008 |
| 456 |
AT4G14440 |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.808 |
0.026 |
| 457 |
AT5G64000 |
AT5G64000
|
ATSAL2, SAL2 |
-0.807 |
0.068 |
| 458 |
AT1G19715 |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.807 |
-0.025 |
| 459 |
AT3G61530 |
AT3G61530
|
PANB2 |
-0.800 |
-0.043 |
| 460 |
AT1G28330 |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.798 |
-0.024 |
| 461 |
AT2G03500 |
AT2G03500
|
[Homeodomain-like superfamily protein] |
-0.791 |
0.001 |
| 462 |
AT5G05250 |
AT5G05250
|
unknown |
-0.790 |
-0.170 |
| 463 |
AT5G43780 |
AT5G43780
|
APS4 |
-0.788 |
-0.025 |
| 464 |
AT1G74460 |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.783 |
-0.087 |
| 465 |
AT5G52640 |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.781 |
-0.006 |
| 466 |
AT2G18360 |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.780 |
0.016 |
| 467 |
AT1G80340 |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
-0.779 |
-0.018 |
| 468 |
AT2G35010 |
AT2G35010
|
ATO1, thioredoxin O1, TO1, thioredoxin O1 |
-0.776 |
0.015 |
| 469 |
AT5G54770 |
AT5G54770
|
THI1, THI4, THIAMINE4, TZ, THIAZOLE REQUIRING |
-0.776 |
-0.001 |
| 470 |
AT5G24170 |
AT5G24170
|
[Got1/Sft2-like vescicle transport protein family] |
-0.776 |
-0.007 |
| 471 |
AT1G74660 |
AT1G74660
|
MIF1, mini zinc finger 1 |
-0.773 |
0.046 |
| 472 |
AT5G62720 |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.773 |
0.015 |
| 473 |
AT5G07650 |
AT5G07650
|
[Actin-binding FH2 protein] |
-0.768 |
no data |
| 474 |
AT1G08210 |
AT1G08210
|
[Eukaryotic aspartyl protease family protein] |
-0.768 |
-0.019 |
| 475 |
AT1G70950 |
AT1G70950
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.766 |
0.054 |
| 476 |
AT1G01610 |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
-0.764 |
-0.053 |
| 477 |
AT2G39730 |
AT2G39730
|
RCA, rubisco activase |
-0.762 |
-0.002 |
| 478 |
AT4G04925 |
AT4G04925
|
unknown |
-0.759 |
no data |
| 479 |
AT5G37690 |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.757 |
-0.046 |
| 480 |
AT4G33220 |
AT4G33220
|
ATPME44, A. THALIANA PECTIN METHYLESTERASE 44, PME44, pectin methylesterase 44 |
-0.755 |
-0.031 |
| 481 |
AT1G27921 |
AT1G27921
|
other RNA |
-0.752 |
no data |
| 482 |
AT5G02380 |
AT5G02380
|
MT2B, metallothionein 2B |
-0.752 |
0.012 |
| 483 |
AT3G49760 |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.751 |
-0.037 |
| 484 |
AT3G15353 |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
-0.744 |
0.010 |