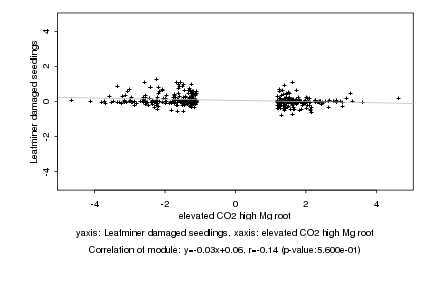
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
elevated CO2 high Mg root |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
AT1G19050 |
AT1G19050
|
ARR7, response regulator 7 |
4.605 |
0.198 |
| 2 |
AT3G43154 |
AT3G43154
|
[pseudogene] |
4.234 |
no data |
| 3 |
AT5G32161 |
AT5G32161
|
unknown |
3.844 |
no data |
| 4 |
AT5G14070 |
AT5G14070
|
ROXY2 |
3.601 |
-0.033 |
| 5 |
AT5G31787 |
AT5G31787
|
unknown |
3.547 |
no data |
| 6 |
AT1G02230 |
AT1G02230
|
ANAC004, NAC domain containing protein 4, NAC004, NAC domain containing protein 4 |
3.433 |
no data |
| 7 |
AT5G32143 |
AT5G32143
|
[pseudogene] |
3.433 |
no data |
| 8 |
AT1G36305 |
AT1G36305
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
3.372 |
no data |
| 9 |
AT5G23030 |
AT5G23030
|
TET12, tetraspanin12 |
3.309 |
0.006 |
| 10 |
AT5G24190 |
AT5G24190
|
[Lipase class 3-related protein] |
3.309 |
no data |
| 11 |
AT5G15240 |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
3.242 |
0.485 |
| 12 |
AT5G26236 |
AT5G26236
|
[gypsy-like retrotransposon family, has a 3.5e-307 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
3.242 |
no data |
| 13 |
AT3G42658 |
AT3G42658
|
SADHU3-2, sadhu non-coding retrotransposon 3-2 |
3.173 |
no data |
| 14 |
AT3G48100 |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
3.143 |
0.221 |
| 15 |
AT4G22485 |
AT4G22485
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
3.071 |
no data |
| 16 |
AT5G26320 |
AT5G26320
|
[TRAF-like family protein] |
3.059 |
no data |
| 17 |
AT2G22290 |
AT2G22290
|
ATRAB-H1D, ARABIDOPSIS RAB GTPASE HOMOLOG H1D, ATRAB6, ARABIDOPSIS RAB GTPASE HOMOLOG 6, ATRABH1D, RAB GTPase homolog H1D, RAB-H1D, RAB GTPASE HOMOLOG H1D, RABH1d, RAB GTPase homolog H1D |
3.023 |
-0.014 |
| 18 |
AT5G25130 |
AT5G25130
|
CYP71B12, cytochrome P450, family 71, subfamily B, polypeptide 12 |
3.017 |
-0.235 |
| 19 |
AT1G21520 |
AT1G21520
|
unknown |
3.017 |
no data |
| 20 |
AT4G19760 |
AT4G19760
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
2.968 |
no data |
| 21 |
AT3G60280 |
AT3G60280
|
UCC3, uclacyanin 3 |
2.961 |
0.027 |
| 22 |
AT5G62380 |
AT5G62380
|
ANAC101, NAC-domain protein 101, NAC101, NAC-domain protein 101, VND6, VASCULAR-RELATED NAC-DOMAIN 6 |
2.868 |
0.017 |
| 23 |
AT2G17890 |
AT2G17890
|
CPK16, calcium-dependent protein kinase 16 |
2.853 |
-0.048 |
| 24 |
AT1G76210 |
AT1G76210
|
unknown |
2.842 |
0.089 |
| 25 |
AT5G63270 |
AT5G63270
|
[RPM1-interacting protein 4 (RIN4) family protein] |
2.841 |
no data |
| 26 |
AT5G28715 |
AT5G28715
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
2.786 |
no data |
| 27 |
AT3G53770 |
AT3G53770
|
[late embryogenesis abundant 3 (LEA3) family protein] |
2.778 |
0.023 |
| 28 |
AT5G07570 |
AT5G07570
|
[glycine/proline-rich protein] |
2.767 |
0.022 |
| 29 |
AT2G44220 |
AT2G44220
|
unknown |
2.750 |
no data |
| 30 |
AT1G23160 |
AT1G23160
|
[Auxin-responsive GH3 family protein] |
2.654 |
0.078 |
| 31 |
AT4G19750 |
AT4G19750
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
2.643 |
no data |
| 32 |
AT5G54530 |
AT5G54530
|
unknown |
2.642 |
-0.315 |
| 33 |
AT1G07160 |
AT1G07160
|
[Protein phosphatase 2C family protein] |
2.642 |
0.024 |
| 34 |
AT4G01580 |
AT4G01580
|
[AP2/B3-like transcriptional factor family protein] |
2.630 |
no data |
| 35 |
AT1G50055 |
AT1G50055
|
TAS1B, trans-acting siRNA1B |
2.574 |
no data |
| 36 |
AT5G43403 |
AT5G43403
|
other RNA |
2.555 |
no data |
| 37 |
AT3G05880 |
AT3G05880
|
RCI2A, RARE-COLD-INDUCIBLE 2A |
2.539 |
0.050 |
| 38 |
AT2G29010 |
AT2G29010
|
[pseudogene] |
2.529 |
no data |
| 39 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
2.492 |
no data |
| 40 |
AT3G29780 |
AT3G29780
|
RALFL27, ralf-like 27 |
2.468 |
-0.042 |
| 41 |
AT1G73810 |
AT1G73810
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
2.457 |
no data |
| 42 |
AT4G25220 |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
2.457 |
-0.058 |
| 43 |
AT2G21400 |
AT2G21400
|
SRS3, SHI-related sequence3 |
2.455 |
no data |
| 44 |
AT2G03821 |
AT2G03821
|
unknown |
2.455 |
no data |
| 45 |
AT3G48740 |
AT3G48740
|
AtSWEET11, SWEET11 |
2.429 |
-0.126 |
| 46 |
AT5G65030 |
AT5G65030
|
unknown |
2.421 |
no data |
| 47 |
AT2G19150 |
AT2G19150
|
[Pectin lyase-like superfamily protein] |
2.414 |
0.003 |
| 48 |
AT5G62920 |
AT5G62920
|
ARR6, response regulator 6 |
2.408 |
0.044 |
| 49 |
AT5G57625 |
AT5G57625
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
2.363 |
no data |
| 50 |
AT4G28850 |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
2.363 |
-0.034 |
| 51 |
AT5G05840 |
AT5G05840
|
unknown |
2.359 |
0.052 |
| 52 |
AT1G08430 |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
2.340 |
-0.019 |
| 53 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
2.325 |
no data |
| 54 |
AT1G48220 |
AT1G48220
|
[Protein kinase superfamily protein] |
2.325 |
-0.035 |
| 55 |
AT5G39250 |
AT5G39250
|
[F-box family protein] |
2.271 |
0.111 |
| 56 |
AT5G25450 |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
2.261 |
0.060 |
| 57 |
AT5G39260 |
AT5G39260
|
ATEXP21, ATEXPA21, expansin A21, ATHEXP ALPHA 1.20, EXP21, EXPANSIN 21, EXPA21, expansin A21 |
2.256 |
-0.043 |
| 58 |
AT5G45240 |
AT5G45240
|
[Disease resistance protein (TIR-NBS-LRR class)] |
2.252 |
0.017 |
| 59 |
AT3G49330 |
AT3G49330
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
2.246 |
no data |
| 60 |
AT3G17180 |
AT3G17180
|
scpl33, serine carboxypeptidase-like 33 |
2.221 |
0.049 |
| 61 |
AT1G51870 |
AT1G51870
|
[protein kinase family protein] |
2.168 |
no data |
| 62 |
AT4G21760 |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
2.156 |
-0.612 |
| 63 |
AT3G03910 |
AT3G03910
|
glutamate dehydrogenase 3 |
2.150 |
-0.287 |
| 64 |
AT4G39770 |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
2.128 |
-0.414 |
| 65 |
AT1G52347 |
AT1G52347
|
other RNA |
2.124 |
no data |
| 66 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
2.112 |
-0.503 |
| 67 |
AT3G07070 |
AT3G07070
|
[Protein kinase superfamily protein] |
2.106 |
-0.042 |
| 68 |
AT5G62420 |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
2.103 |
-0.005 |
| 69 |
AT4G30993 |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
2.101 |
-0.186 |
| 70 |
AT1G48325 |
AT1G48325
|
[Expressed protein] |
2.098 |
no data |
| 71 |
AT5G48850 |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
2.093 |
-0.023 |
| 72 |
AT1G49500 |
AT1G49500
|
unknown |
2.084 |
0.187 |
| 73 |
AT5G24040 |
AT5G24040
|
unknown |
2.074 |
no data |
| 74 |
AT5G66658 |
AT5G66658
|
unknown |
2.073 |
no data |
| 75 |
AT1G49100 |
AT1G49100
|
[Leucine-rich repeat protein kinase family protein] |
2.064 |
0.006 |
| 76 |
AT1G68430 |
AT1G68430
|
unknown |
2.042 |
-0.170 |
| 77 |
AT2G16790 |
AT2G16790
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
2.025 |
0.204 |
| 78 |
AT1G21670 |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
2.013 |
0.247 |
| 79 |
AT2G26150 |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
2.011 |
-0.371 |
| 80 |
AT5G41560 |
AT5G41560
|
unknown |
1.989 |
0.087 |
| 81 |
AT1G16530 |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
1.987 |
-0.047 |
| 82 |
AT1G51840 |
AT1G51840
|
[protein kinase-related] |
1.975 |
-0.021 |
| 83 |
AT3G48940 |
AT3G48940
|
[Remorin family protein] |
1.974 |
-0.069 |
| 84 |
AT2G20520 |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
1.970 |
0.040 |
| 85 |
AT3G53010 |
AT3G53010
|
unknown |
1.970 |
no data |
| 86 |
AT5G26130 |
AT5G26130
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.958 |
-0.020 |
| 87 |
AT5G24920 |
AT5G24920
|
AtGDU5, glutamine dumper 5, GDU5, glutamine dumper 5 |
1.946 |
no data |
| 88 |
AT4G30670 |
AT4G30670
|
[Putative membrane lipoprotein] |
1.945 |
0.014 |
| 89 |
AT1G70890 |
AT1G70890
|
MLP43, MLP-like protein 43 |
1.933 |
-0.091 |
| 90 |
AT4G31330 |
AT4G31330
|
unknown |
1.922 |
-0.018 |
| 91 |
AT1G06120 |
AT1G06120
|
[Fatty acid desaturase family protein] |
1.919 |
no data |
| 92 |
AT1G10470 |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
1.915 |
-0.061 |
| 93 |
AT3G59730 |
AT3G59730
|
LecRK-V.6, L-type lectin receptor kinase V.6 |
1.913 |
-0.005 |
| 94 |
AT5G03890 |
AT5G03890
|
unknown |
1.907 |
no data |
| 95 |
AT4G24090 |
AT4G24090
|
unknown |
1.897 |
-0.195 |
| 96 |
AT3G09925 |
AT3G09925
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.880 |
no data |
| 97 |
AT1G58037 |
AT1G58037
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.877 |
no data |
| 98 |
AT1G51830 |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
1.865 |
0.017 |
| 99 |
AT3G62450 |
AT3G62450
|
unknown |
1.861 |
0.050 |
| 100 |
AT2G29620 |
AT2G29620
|
unknown |
1.856 |
-0.016 |
| 101 |
AT1G19900 |
AT1G19900
|
[glyoxal oxidase-related protein] |
1.855 |
-0.012 |
| 102 |
AT5G21100 |
AT5G21100
|
[Plant L-ascorbate oxidase] |
1.848 |
-0.427 |
| 103 |
AT5G56670 |
AT5G56670
|
[Ribosomal protein S30 family protein] |
1.832 |
0.016 |
| 104 |
AT1G23090 |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
1.829 |
0.044 |
| 105 |
AT1G14960 |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.829 |
-0.003 |
| 106 |
AT5G58360 |
AT5G58360
|
ATOFP3, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 3, OFP3, ovate family protein 3 |
1.827 |
-0.048 |
| 107 |
AT4G22610 |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.825 |
0.029 |
| 108 |
AT2G11810 |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
1.805 |
-0.475 |
| 109 |
AT1G29465 |
AT1G29465
|
unknown |
1.802 |
no data |
| 110 |
AT1G08770 |
AT1G08770
|
PRA1.E, prenylated RAB acceptor 1.E |
1.802 |
0.102 |
| 111 |
AT3G12040 |
AT3G12040
|
[DNA-3-methyladenine glycosylase (MAG)] |
1.788 |
0.230 |
| 112 |
AT1G07690 |
AT1G07690
|
unknown |
1.779 |
-0.006 |
| 113 |
AT3G46760 |
AT3G46760
|
LecRK-S.3, L-type lectin receptor kinase S.3 |
1.755 |
-0.050 |
| 114 |
AT5G40730 |
AT5G40730
|
AGP24, arabinogalactan protein 24, ATAGP24, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 24 |
1.728 |
-0.027 |
| 115 |
AT4G16000 |
AT4G16000
|
unknown |
1.727 |
0.661 |
| 116 |
AT5G07000 |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
1.727 |
-0.129 |
| 117 |
AT3G05935 |
AT3G05935
|
unknown |
1.725 |
no data |
| 118 |
AT2G15970 |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
1.720 |
0.124 |
| 119 |
AT1G53887 |
AT1G53887
|
unknown |
1.713 |
no data |
| 120 |
AT4G22680 |
AT4G22680
|
AtMYB85, myb domain protein 85, MYB85, myb domain protein 85 |
1.712 |
0.121 |
| 121 |
AT4G19950 |
AT4G19950
|
unknown |
1.708 |
no data |
| 122 |
AT2G28270 |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.701 |
-0.014 |
| 123 |
AT2G25240 |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
1.694 |
0.064 |
| 124 |
AT3G53360 |
AT3G53360
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
1.683 |
-0.082 |
| 125 |
AT4G08380 |
AT4G08380
|
[Proline-rich extensin-like family protein] |
1.673 |
-0.001 |
| 126 |
AT1G73630 |
AT1G73630
|
[EF hand calcium-binding protein family] |
1.672 |
-0.046 |
| 127 |
AT4G28620 |
AT4G28620
|
ABCB24, ATP-binding cassette B24, ATATM2, ARABIDOPSIS THALIANA ABC TRANSPORTER OF THE MITOCHONDRION 2, ATM2, ABC transporter of the mitochondrion 2 |
1.664 |
no data |
| 128 |
AT5G40942 |
AT5G40942
|
RNR2B, ribonucleotide reductase 2B |
1.664 |
no data |
| 129 |
AT1G67370 |
AT1G67370
|
ASY1, ASYNAPTIC 1, ATASY1 |
1.657 |
-0.015 |
| 130 |
AT5G01740 |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
1.655 |
-0.425 |
| 131 |
AT1G11655 |
AT1G11655
|
unknown |
1.646 |
no data |
| 132 |
AT4G05590 |
AT4G05590
|
unknown |
1.644 |
0.170 |
| 133 |
AT1G04240 |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
1.642 |
-0.321 |
| 134 |
AT5G13930 |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
1.639 |
-0.402 |
| 135 |
AT1G80800 |
AT1G80800
|
[pseudogene] |
1.635 |
-0.041 |
| 136 |
AT5G06330 |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.633 |
-0.067 |
| 137 |
AT4G36240 |
AT4G36240
|
GATA7, GATA transcription factor 7 |
1.632 |
no data |
| 138 |
AT3G48920 |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
1.621 |
0.061 |
| 139 |
AT3G28345 |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
1.620 |
-0.168 |
| 140 |
AT1G64380 |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
1.614 |
1.093 |
| 141 |
AT5G03995 |
AT5G03995
|
unknown |
1.609 |
no data |
| 142 |
AT5G40980 |
AT5G40980
|
unknown |
1.602 |
-0.173 |
| 143 |
AT5G05270 |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
1.600 |
-0.730 |
| 144 |
AT5G24270 |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
1.597 |
0.164 |
| 145 |
AT2G25160 |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
1.597 |
0.025 |
| 146 |
AT5G20500 |
AT5G20500
|
[Glutaredoxin family protein] |
1.585 |
0.077 |
| 147 |
AT2G41780 |
AT2G41780
|
unknown |
1.583 |
0.153 |
| 148 |
AT3G27027 |
AT3G27027
|
unknown |
1.582 |
no data |
| 149 |
AT3G57870 |
AT3G57870
|
AHUS5, ATSCE1, SUMO CONJUGATION ENZYME 1, EMB1637, EMBRYO DEFECTIVE 1637, SCE1, sumo conjugation enzyme 1, SCE1A, SUMO CONJUGATING ENZYME 1A |
1.574 |
0.068 |
| 150 |
AT5G45810 |
AT5G45810
|
CIPK19, CBL-interacting protein kinase 19, SnRK3.5, SNF1-RELATED PROTEIN KINASE 3.5 |
1.572 |
0.110 |
| 151 |
AT1G48780 |
AT1G48780
|
unknown |
1.571 |
-0.100 |
| 152 |
AT2G32740 |
AT2G32740
|
ATGT13, ARABIDOPSIS GALACTOSYLTRANSFERASE 13, GT13, galactosyltransferase 13 |
1.547 |
0.074 |
| 153 |
AT5G44130 |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
1.539 |
-0.437 |
| 154 |
AT3G18773 |
AT3G18773
|
[RING/U-box superfamily protein] |
1.534 |
no data |
| 155 |
AT4G04810 |
AT4G04810
|
ATMSRB4, methionine sulfoxide reductase B4, MSRB4, methionine sulfoxide reductase B4 |
1.534 |
no data |
| 156 |
AT5G50760 |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
1.534 |
0.491 |
| 157 |
AT1G59730 |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
1.527 |
-0.044 |
| 158 |
AT1G19240 |
AT1G19240
|
unknown |
1.524 |
0.131 |
| 159 |
AT5G46900 |
AT5G46900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.519 |
no data |
| 160 |
AT3G28360 |
AT3G28360
|
ABCB16, ATP-binding cassette B16, PGP16, P-glycoprotein 16 |
1.517 |
0.027 |
| 161 |
AT3G53980 |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.517 |
0.048 |
| 162 |
AT1G51880 |
AT1G51880
|
RHS6, root hair specific 6 |
1.514 |
0.012 |
| 163 |
AT4G25820 |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
1.511 |
0.042 |
| 164 |
AT1G53560 |
AT1G53560
|
[Ribosomal protein L18ae family] |
1.510 |
0.273 |
| 165 |
AT2G37460 |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
1.502 |
-0.227 |
| 166 |
AT2G34180 |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
1.499 |
-0.028 |
| 167 |
AT3G01080 |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
1.495 |
0.225 |
| 168 |
AT3G05932 |
AT3G05932
|
|
1.494 |
no data |
| 169 |
AT2G15370 |
AT2G15370
|
ATFUT5, ARABIDOPSIS FUCOSYLTRANSFERASE 5, FUT5, fucosyltransferase 5 |
1.493 |
no data |
| 170 |
AT2G38760 |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
1.488 |
0.531 |
| 171 |
AT1G51430 |
AT1G51430
|
unknown |
1.486 |
-0.003 |
| 172 |
AT4G33800 |
AT4G33800
|
unknown |
1.480 |
-0.036 |
| 173 |
AT1G54950 |
AT1G54950
|
unknown |
1.477 |
no data |
| 174 |
AT4G02270 |
AT4G02270
|
RHS13, root hair specific 13 |
1.475 |
0.023 |
| 175 |
AT5G19800 |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
1.474 |
0.012 |
| 176 |
AT1G72870 |
AT1G72870
|
[Disease resistance protein (TIR-NBS class)] |
1.473 |
0.047 |
| 177 |
AT4G37560 |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
1.473 |
-0.288 |
| 178 |
AT1G10480 |
AT1G10480
|
ZFP5, zinc finger protein 5 |
1.472 |
0.016 |
| 179 |
AT1G69240 |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
1.471 |
-0.045 |
| 180 |
AT1G62975 |
AT1G62975
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.469 |
no data |
| 181 |
AT1G14630 |
AT1G14630
|
unknown |
1.469 |
no data |
| 182 |
AT4G17340 |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
1.458 |
0.103 |
| 183 |
AT1G61080 |
AT1G61080
|
[Hydroxyproline-rich glycoprotein family protein] |
1.456 |
-0.001 |
| 184 |
AT2G41178 |
AT2G41178
|
other RNA |
1.455 |
no data |
| 185 |
AT1G15460 |
AT1G15460
|
ATBOR4, ARABIDOPSIS THALIANA REQUIRES HIGH BORON 4, BOR4, REQUIRES HIGH BORON 4 |
1.455 |
-0.037 |
| 186 |
AT5G38100 |
AT5G38100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.448 |
no data |
| 187 |
AT1G03850 |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
1.445 |
0.483 |
| 188 |
AT1G59590 |
AT1G59590
|
ZCF37 |
1.438 |
0.231 |
| 189 |
AT4G29905 |
AT4G29905
|
unknown |
1.434 |
no data |
| 190 |
AT4G00680 |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
1.433 |
-0.109 |
| 191 |
AT1G33280 |
AT1G33280
|
ANAC015, NAC domain containing protein 15, BRN1, BEARSKIN 1, NAC015, NAC domain containing protein 15 |
1.426 |
0.024 |
| 192 |
AT3G05800 |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
1.425 |
0.447 |
| 193 |
AT4G20780 |
AT4G20780
|
CML42, calmodulin like 42 |
1.421 |
0.186 |
| 194 |
AT4G38830 |
AT4G38830
|
CRK26, cysteine-rich RLK (RECEPTOR-like protein kinase) 26 |
1.420 |
0.106 |
| 195 |
AT2G27370 |
AT2G27370
|
CASP3, Casparian strip membrane domain protein 3 |
1.418 |
0.047 |
| 196 |
AT2G34340 |
AT2G34340
|
unknown |
1.416 |
-0.193 |
| 197 |
AT3G47790 |
AT3G47790
|
ABCA8, ATP-binding cassette A8, ATATH7, ATH7, ABC2 homolog 7 |
1.416 |
no data |
| 198 |
AT2G21830 |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.415 |
-0.343 |
| 199 |
AT1G54970 |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
1.412 |
0.010 |
| 200 |
AT3G46270 |
AT3G46270
|
[receptor protein kinase-related] |
1.412 |
-0.003 |
| 201 |
AT1G12560 |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
1.410 |
-0.057 |
| 202 |
AT2G38600 |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
1.405 |
-0.016 |
| 203 |
AT1G70895 |
AT1G70895
|
CLE17, CLAVATA3/ESR-RELATED 17 |
1.403 |
-0.087 |
| 204 |
AT1G75750 |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
1.400 |
-0.394 |
| 205 |
AT1G09140 |
AT1G09140
|
At-SR30, Serine/Arginine-Rich Protein Splicing Factor 30, ATSRP30, SERINE-ARGININE PROTEIN 30, ATSRP30.1, ATSRP30.2, SR30, Serine/Arginine-Rich Protein Splicing Factor 30 |
1.399 |
-0.171 |
| 206 |
AT5G38790 |
AT5G38790
|
unknown |
1.398 |
-0.018 |
| 207 |
AT1G54095 |
AT1G54095
|
unknown |
1.398 |
no data |
| 208 |
AT5G23950 |
AT5G23950
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
1.394 |
no data |
| 209 |
AT5G47990 |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
1.390 |
0.026 |
| 210 |
AT5G47450 |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
1.389 |
0.050 |
| 211 |
AT4G34800 |
AT4G34800
|
SAUR4, SMALL AUXIN UPREGULATED RNA 4 |
1.388 |
no data |
| 212 |
AT5G55510 |
AT5G55510
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
1.388 |
no data |
| 213 |
AT1G33910 |
AT1G33910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.387 |
-0.020 |
| 214 |
AT2G46250 |
AT2G46250
|
[myosin heavy chain-related] |
1.381 |
-0.486 |
| 215 |
AT2G32140 |
AT2G32140
|
[transmembrane receptors] |
1.381 |
0.929 |
| 216 |
AT3G12580 |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
1.380 |
-0.228 |
| 217 |
AT4G26220 |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
1.379 |
-0.087 |
| 218 |
AT3G49960 |
AT3G49960
|
[Peroxidase superfamily protein] |
1.378 |
-0.057 |
| 219 |
AT5G17210 |
AT5G17210
|
unknown |
1.374 |
0.193 |
| 220 |
AT1G68760 |
AT1G68760
|
ATNUDT1, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 1, ATNUDX1, nudix hydrolase 1, NUDX1, nudix hydrolase 1, NUDX1, NUDIX HYDROLASE 1 |
1.372 |
-0.037 |
| 221 |
AT4G36945 |
AT4G36945
|
[PLC-like phosphodiesterases superfamily protein] |
1.371 |
no data |
| 222 |
AT5G48840 |
AT5G48840
|
ATPTS, ARABIDOPSIS THALIANA PANTOTHENATE SYNTHETASE, PANC, homolog of bacterial PANC, PTS, PANTOTHENATE SYNTHETASE |
1.371 |
-0.063 |
| 223 |
AT4G02850 |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
1.370 |
-0.204 |
| 224 |
AT4G38140 |
AT4G38140
|
[RING/U-box superfamily protein] |
1.367 |
no data |
| 225 |
AT2G27035 |
AT2G27035
|
AtENODL20, ENODL20, early nodulin-like protein 20 |
1.362 |
no data |
| 226 |
AT2G20030 |
AT2G20030
|
[RING/U-box superfamily protein] |
1.362 |
0.080 |
| 227 |
AT2G31150 |
AT2G31150
|
[ATP binding] |
1.359 |
no data |
| 228 |
AT2G38750 |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
1.354 |
0.422 |
| 229 |
AT1G33817 |
AT1G33817
|
[copia-like retrotransposon family, has a 8.1e-100 P-value blast match to gb|AAO73521.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
1.353 |
0.029 |
| 230 |
AT4G12330 |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
1.351 |
0.103 |
| 231 |
AT2G19780 |
AT2G19780
|
[Leucine-rich repeat (LRR) family protein] |
1.348 |
0.045 |
| 232 |
AT2G41510 |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
1.344 |
0.046 |
| 233 |
AT1G05900 |
AT1G05900
|
ATNTH2, NTH2, endonuclease III 2 |
1.344 |
-0.111 |
| 234 |
AT1G65310 |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
1.343 |
-0.381 |
| 235 |
AT5G58010 |
AT5G58010
|
LRL3, LJRHL1-like 3 |
1.342 |
-0.076 |
| 236 |
AT4G25400 |
AT4G25400
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.337 |
no data |
| 237 |
AT1G73875 |
AT1G73875
|
[DNAse I-like superfamily protein] |
1.335 |
no data |
| 238 |
AT1G50170 |
AT1G50170
|
ATSIRB, sirohydrochlorin ferrochelatase B, SIRB, sirohydrochlorin ferrochelatase B |
1.326 |
-0.026 |
| 239 |
AT3G22570 |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.322 |
0.074 |
| 240 |
AT3G25495 |
AT3G25495
|
[expressed protein] |
1.318 |
no data |
| 241 |
AT4G26560 |
AT4G26560
|
CBL7, calcineurin B-like protein 7 |
1.314 |
-0.003 |
| 242 |
AT1G76250 |
AT1G76250
|
unknown |
1.314 |
-0.007 |
| 243 |
AT1G48740 |
AT1G48740
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.313 |
no data |
| 244 |
AT2G20080 |
AT2G20080
|
TIE2, TCP Interactor containing EAR motif protein 2 |
1.310 |
no data |
| 245 |
AT4G29740 |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
1.310 |
0.661 |
| 246 |
AT5G06450 |
AT5G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
1.309 |
no data |
| 247 |
AT1G52660 |
AT1G52660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.309 |
0.028 |
| 248 |
AT1G35515 |
AT1G35515
|
HOS10, high response to osmotic stress 10, MYB8 |
1.301 |
0.015 |
| 249 |
AT3G05160 |
AT3G05160
|
[Major facilitator superfamily protein] |
1.300 |
no data |
| 250 |
AT4G30320 |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.298 |
-0.030 |
| 251 |
AT1G02475 |
AT1G02475
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.298 |
-0.089 |
| 252 |
AT2G43590 |
AT2G43590
|
[Chitinase family protein] |
1.297 |
0.393 |
| 253 |
AT4G24450 |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
1.297 |
-0.082 |
| 254 |
AT2G17975 |
AT2G17975
|
[zinc finger (Ran-binding) family protein] |
1.295 |
0.044 |
| 255 |
AT1G18290 |
AT1G18290
|
unknown |
1.292 |
0.025 |
| 256 |
AT5G24165 |
AT5G24165
|
unknown |
1.292 |
-0.203 |
| 257 |
AT3G01750 |
AT3G01750
|
[Ankyrin repeat family protein] |
1.284 |
0.054 |
| 258 |
AT5G08640 |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
1.281 |
-0.745 |
| 259 |
AT1G67110 |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
1.277 |
-0.062 |
| 260 |
AT2G17830 |
AT2G17830
|
[F-box and associated interaction domains-containing protein] |
1.271 |
no data |
| 261 |
AT1G33813 |
AT1G33813
|
[copia-like retrotransposon family, has a 5.8e-39 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
1.271 |
-0.014 |
| 262 |
AT4G39361 |
AT4G39361
|
[snoRNA] |
1.268 |
no data |
| 263 |
AT4G27480 |
AT4G27480
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
1.267 |
0.091 |
| 264 |
AT5G67400 |
AT5G67400
|
RHS19, root hair specific 19 |
1.266 |
-0.057 |
| 265 |
AT1G13080 |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
1.259 |
-0.343 |
| 266 |
AT5G05500 |
AT5G05500
|
MOP10 |
1.258 |
0.001 |
| 267 |
AT3G07274 |
AT3G07274
|
|
1.257 |
0.198 |
| 268 |
AT2G35000 |
AT2G35000
|
ATL9, Arabidopsis toxicos en levadura 9 |
1.251 |
0.194 |
| 269 |
AT5G40630 |
AT5G40630
|
[Ubiquitin-like superfamily protein] |
1.244 |
-0.090 |
| 270 |
AT5G26300 |
AT5G26300
|
[TRAF-like family protein] |
1.243 |
no data |
| 271 |
AT3G43740 |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
1.243 |
0.048 |
| 272 |
AT3G04020 |
AT3G04020
|
unknown |
1.242 |
0.047 |
| 273 |
AT1G68360 |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
1.240 |
0.086 |
| 274 |
AT1G05250 |
AT1G05250
|
AtPRX1, PRX2, peroxidase 2 |
1.239 |
no data |
| 275 |
AT2G41380 |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.239 |
0.623 |
| 276 |
AT3G44510 |
AT3G44510
|
[alpha/beta-Hydrolases superfamily protein] |
1.238 |
-0.350 |
| 277 |
AT1G30757 |
AT1G30757
|
unknown |
1.236 |
no data |
| 278 |
AT1G76590 |
AT1G76590
|
[PLATZ transcription factor family protein] |
1.234 |
0.701 |
| 279 |
AT2G22190 |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
1.231 |
-0.000 |
| 280 |
AT1G05240 |
AT1G05240
|
[Peroxidase superfamily protein] |
1.230 |
no data |
| 281 |
AT5G47630 |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
1.222 |
-0.189 |
| 282 |
AT1G18980 |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
1.217 |
-0.001 |
| 283 |
AT4G29530 |
AT4G29530
|
[Pyridoxal phosphate phosphatase-related protein] |
1.214 |
0.046 |
| 284 |
AT4G33880 |
AT4G33880
|
RSL2, ROOT HAIR DEFECTIVE 6-LIKE 2 |
1.213 |
no data |
| 285 |
AT1G10682 |
AT1G10682
|
other RNA |
1.211 |
no data |
| 286 |
AT2G22122 |
AT2G22122
|
unknown |
1.210 |
no data |
| 287 |
AT2G39705 |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
1.206 |
-0.376 |
| 288 |
AT4G02970 |
AT4G02970
|
AT7SL-1, 7SL RNA1 |
1.205 |
no data |
| 289 |
AT5G48630 |
AT5G48630
|
[Cyclin family protein] |
1.205 |
-0.095 |
| 290 |
AT3G50840 |
AT3G50840
|
[Phototropic-responsive NPH3 family protein] |
1.203 |
no data |
| 291 |
AT1G70860 |
AT1G70860
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.202 |
no data |
| 292 |
AT1G22065 |
AT1G22065
|
unknown |
1.197 |
no data |
| 293 |
AT2G29995 |
AT2G29995
|
unknown |
1.194 |
0.308 |
| 294 |
AT1G54050 |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
1.192 |
-0.358 |
| 295 |
AT1G31320 |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
1.190 |
-0.224 |
| 296 |
AT4G15800 |
AT4G15800
|
RALFL33, ralf-like 33 |
1.188 |
0.018 |
| 297 |
AT5G23955 |
AT5G23955
|
[non-LTR retrotransposon family (LINE), has a 2.6e-41 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
1.187 |
no data |
| 298 |
AT3G21340 |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
1.186 |
-0.023 |
| 299 |
AT1G15100 |
AT1G15100
|
RHA2A, RING-H2 finger A2A |
1.185 |
0.066 |
| 300 |
AT3G52760 |
AT3G52760
|
[Integral membrane Yip1 family protein] |
1.181 |
0.034 |
| 301 |
AT2G47520 |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-4.676 |
0.097 |
| 302 |
AT1G06030 |
AT1G06030
|
[pfkB-like carbohydrate kinase family protein] |
-4.453 |
no data |
| 303 |
AT2G41810 |
AT2G41810
|
unknown |
-4.122 |
0.055 |
| 304 |
AT5G24550 |
AT5G24550
|
BGLU32, beta glucosidase 32 |
-3.828 |
-0.025 |
| 305 |
AT1G47395 |
AT1G47395
|
unknown |
-3.731 |
no data |
| 306 |
AT1G58300 |
AT1G58300
|
HO4, heme oxygenase 4 |
-3.723 |
0.054 |
| 307 |
AT5G59170 |
AT5G59170
|
[Proline-rich extensin-like family protein] |
-3.698 |
-0.069 |
| 308 |
AT1G19210 |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-3.595 |
0.304 |
| 309 |
AT2G17850 |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-3.542 |
-0.025 |
| 310 |
AT2G17845 |
AT2G17845
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-3.486 |
0.026 |
| 311 |
AT3G19025 |
AT3G19025
|
[pseudogene] |
-3.483 |
no data |
| 312 |
AT1G45140 |
AT1G45140
|
[non-LTR retrotransposon family (LINE), has a 1.4e-37 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-3.427 |
no data |
| 313 |
AT5G56620 |
AT5G56620
|
anac099, NAC domain containing protein 99, NAC099, NAC domain containing protein 99 |
-3.366 |
-0.030 |
| 314 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
-3.351 |
0.866 |
| 315 |
AT4G33070 |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-3.315 |
-0.015 |
| 316 |
AT1G52120 |
AT1G52120
|
[Mannose-binding lectin superfamily protein] |
-3.303 |
no data |
| 317 |
AT4G37050 |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
-3.237 |
-0.094 |
| 318 |
AT4G25490 |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-3.235 |
0.333 |
| 319 |
AT2G36270 |
AT2G36270
|
ABI5, ABA INSENSITIVE 5, AtABI5, GIA1, GROWTH-INSENSITIVITY TO ABA 1 |
-3.167 |
0.033 |
| 320 |
AT5G42290 |
AT5G42290
|
[transcription activator-related] |
-3.167 |
-0.007 |
| 321 |
AT3G14300 |
AT3G14300
|
ATPME26, A. THALIANA PECTIN METHYLESTERASE 26, ATPMEPCRC, PME26, PECTIN METHYLESTERASE 26 |
-3.167 |
0.070 |
| 322 |
AT4G24110 |
AT4G24110
|
unknown |
-3.139 |
0.395 |
| 323 |
AT5G02190 |
AT5G02190
|
ATASP38, ARABIDOPSIS THALIANA ASPARTIC PROTEASE 38, EMB24, EMBRYO DEFECTIVE 24, PCS1, PROMOTION OF CELL SURVIVAL 1 |
-3.094 |
-0.004 |
| 324 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-3.085 |
0.615 |
| 325 |
AT5G54095 |
AT5G54095
|
unknown |
-3.017 |
0.020 |
| 326 |
AT2G47190 |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
-3.013 |
0.729 |
| 327 |
AT5G10040 |
AT5G10040
|
unknown |
-3.008 |
0.076 |
| 328 |
AT4G33560 |
AT4G33560
|
[Wound-responsive family protein] |
-2.979 |
-0.020 |
| 329 |
AT4G29050 |
AT4G29050
|
LecRK-V.9, L-type lectin receptor kinase V.9 |
-2.965 |
0.255 |
| 330 |
AT3G20395 |
AT3G20395
|
[RING/U-box superfamily protein] |
-2.955 |
no data |
| 331 |
AT4G18080 |
AT4G18080
|
unknown |
-2.937 |
-0.047 |
| 332 |
AT5G22240 |
AT5G22240
|
ATOFP10, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 10, OFP10 |
-2.936 |
no data |
| 333 |
AT1G56242 |
AT1G56242
|
other RNA |
-2.922 |
no data |
| 334 |
AT1G56240 |
AT1G56240
|
AtPP2-B13, phloem protein 2-B13, PP2-B13, phloem protein 2-B13 |
-2.922 |
no data |
| 335 |
AT3G22120 |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-2.883 |
-0.224 |
| 336 |
AT3G22121 |
AT3G22121
|
other RNA |
-2.880 |
0.022 |
| 337 |
AT5G42053 |
AT5G42053
|
unknown |
-2.850 |
no data |
| 338 |
AT1G58007 |
AT1G58007
|
unknown |
-2.850 |
no data |
| 339 |
ATCG00530 |
ATCG00530
|
YCF10 |
-2.836 |
-0.066 |
| 340 |
AT2G05441 |
AT2G05441
|
|
-2.795 |
no data |
| 341 |
AT1G42980 |
AT1G42980
|
[Actin-binding FH2 (formin homology 2) family protein] |
-2.773 |
no data |
| 342 |
AT4G01360 |
AT4G01360
|
BPS3, BYPASS 3 |
-2.753 |
no data |
| 343 |
AT2G30670 |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-2.720 |
0.014 |
| 344 |
AT2G30660 |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-2.651 |
0.019 |
| 345 |
AT4G35295 |
AT4G35295
|
[homoserine kinase, putative / HSK, putative] |
-2.637 |
no data |
| 346 |
AT4G27440 |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-2.637 |
0.136 |
| 347 |
AT1G15640 |
AT1G15640
|
unknown |
-2.616 |
-0.010 |
| 348 |
AT3G22910 |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-2.616 |
0.235 |
| 349 |
AT4G37780 |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
-2.596 |
0.048 |
| 350 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-2.591 |
1.097 |
| 351 |
AT5G42146 |
AT5G42146
|
unknown |
-2.571 |
no data |
| 352 |
AT1G69920 |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
-2.570 |
0.280 |
| 353 |
AT5G22530 |
AT5G22530
|
unknown |
-2.568 |
0.375 |
| 354 |
AT5G52680 |
AT5G52680
|
[Copper transport protein family] |
-2.568 |
0.016 |
| 355 |
AT1G09080 |
AT1G09080
|
BIP3, binding protein 3 |
-2.564 |
0.017 |
| 356 |
AT2G22870 |
AT2G22870
|
EMB2001, embryo defective 2001 |
-2.564 |
-0.127 |
| 357 |
AT1G69490 |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-2.561 |
0.011 |
| 358 |
AT2G26400 |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-2.544 |
-0.002 |
| 359 |
AT2G03380 |
AT2G03380
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-2.510 |
-0.028 |
| 360 |
AT4G01780 |
AT4G01780
|
[XH/XS domain-containing protein] |
-2.510 |
no data |
| 361 |
AT3G26060 |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
-2.487 |
-0.201 |
| 362 |
AT4G28790 |
AT4G28790
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-2.487 |
no data |
| 363 |
AT1G77120 |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-2.460 |
0.198 |
| 364 |
AT3G22060 |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-2.449 |
0.194 |
| 365 |
AT1G29930 |
AT1G29930
|
AB140, CAB1, chlorophyll A/B binding protein 1, CAB140, CHLOROPHYLL A/B PROTEIN 140, LHCB1.3, LIGHT-HARVESTING CHLOROPHYLL A/B-PROTEIN 1.3 |
-2.440 |
no data |
| 366 |
AT2G30750 |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-2.424 |
0.852 |
| 367 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-2.419 |
0.837 |
| 368 |
AT3G12230 |
AT3G12230
|
scpl14, serine carboxypeptidase-like 14 |
-2.401 |
no data |
| 369 |
AT5G65205 |
AT5G65205
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-2.397 |
-0.011 |
| 370 |
AT5G39680 |
AT5G39680
|
EMB2744, EMBRYO DEFECTIVE 2744 |
-2.381 |
0.068 |
| 371 |
AT1G16260 |
AT1G16260
|
[Wall-associated kinase family protein] |
-2.374 |
-0.145 |
| 372 |
AT1G43800 |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN-DESATURASE6 |
-2.365 |
0.024 |
| 373 |
AT3G15370 |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-2.358 |
0.003 |
| 374 |
AT5G35790 |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-2.320 |
-0.297 |
| 375 |
AT5G13230 |
AT5G13230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.319 |
-0.008 |
| 376 |
AT1G05650 |
AT1G05650
|
[Pectin lyase-like superfamily protein] |
-2.318 |
no data |
| 377 |
AT1G52130 |
AT1G52130
|
[Mannose-binding lectin superfamily protein] |
-2.277 |
no data |
| 378 |
AT1G59124 |
AT1G59124
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-2.272 |
no data |
| 379 |
AT5G52700 |
AT5G52700
|
[Copper transport protein family] |
-2.266 |
no data |
| 380 |
AT1G65680 |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-2.255 |
-0.059 |
| 381 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
-2.251 |
1.263 |
| 382 |
AT1G32470 |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-2.250 |
-0.189 |
| 383 |
AT3G45120 |
AT3G45120
|
unknown |
-2.250 |
0.012 |
| 384 |
AT2G20540 |
AT2G20540
|
MEF21, mitochondrial editing factor 21 |
-2.250 |
-0.077 |
| 385 |
AT1G29860 |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
-2.249 |
0.080 |
| 386 |
AT1G12805 |
AT1G12805
|
[nucleotide binding] |
-2.246 |
no data |
| 387 |
AT3G26200 |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-2.239 |
0.246 |
| 388 |
AT3G47760 |
AT3G47760
|
ABCA5, ATP-binding cassette A5, ATATH4, A. THALIANA ABC2 HOMOLOG 4, ATH4, ABC2 homolog 4 |
-2.237 |
no data |
| 389 |
AT5G64110 |
AT5G64110
|
[Peroxidase superfamily protein] |
-2.229 |
0.028 |
| 390 |
AT1G14345 |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-2.224 |
-0.416 |
| 391 |
AT3G61220 |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
-2.222 |
-0.106 |
| 392 |
AT3G15520 |
AT3G15520
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-2.206 |
-0.272 |
| 393 |
ATCG00890 |
ATCG00890
|
NDHB.1 |
-2.205 |
no data |
| 394 |
AT2G47950 |
AT2G47950
|
unknown |
-2.205 |
0.499 |
| 395 |
AT1G26420 |
AT1G26420
|
[FAD-binding Berberine family protein] |
-2.194 |
0.840 |
| 396 |
AT5G47220 |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-2.180 |
0.598 |
| 397 |
AT1G33055 |
AT1G33055
|
unknown |
-2.170 |
0.015 |
| 398 |
AT4G28405 |
AT4G28405
|
[Expressed protein] |
-2.133 |
no data |
| 399 |
AT3G47380 |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-2.092 |
0.643 |
| 400 |
AT3G52970 |
AT3G52970
|
CYP76G1, cytochrome P450, family 76, subfamily G, polypeptide 1 |
-2.084 |
-0.039 |
| 401 |
AT1G01560 |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-2.081 |
0.716 |
| 402 |
AT5G59490 |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-2.068 |
0.224 |
| 403 |
AT1G15540 |
AT1G15540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.020 |
no data |
| 404 |
AT1G14550 |
AT1G14550
|
[Peroxidase superfamily protein] |
-2.008 |
0.181 |
| 405 |
AT1G26700 |
AT1G26700
|
ATMLO14, MILDEW RESISTANCE LOCUS O 14, MLO14, MILDEW RESISTANCE LOCUS O 14 |
-2.008 |
0.018 |
| 406 |
AT1G21810 |
AT1G21810
|
unknown |
-2.006 |
-0.076 |
| 407 |
AT5G50590 |
AT5G50590
|
AtHSD4, hydroxysteroid dehydrogenase 4, HSD4, hydroxysteroid dehydrogenase 4 |
-2.004 |
no data |
| 408 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-1.980 |
0.370 |
| 409 |
AT2G33793 |
AT2G33793
|
unknown |
-1.979 |
no data |
| 410 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
-1.978 |
no data |
| 411 |
AT4G39580 |
AT4G39580
|
[Galactose oxidase/kelch repeat superfamily protein] |
-1.959 |
no data |
| 412 |
AT5G61430 |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
-1.959 |
0.002 |
| 413 |
AT2G07765 |
AT2G07765
|
[tRNA-Tyr (anticodon: GTA)] |
-1.953 |
no data |
| 414 |
AT1G18745 |
AT1G18745
|
|
-1.940 |
no data |
| 415 |
AT3G47720 |
AT3G47720
|
SRO4, similar to RCD one 4 |
-1.907 |
no data |
| 416 |
AT3G10040 |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-1.894 |
-0.105 |
| 417 |
AT3G56940 |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
-1.879 |
-0.083 |
| 418 |
AT5G22520 |
AT5G22520
|
unknown |
-1.859 |
no data |
| 419 |
AT1G13610 |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
-1.859 |
-0.034 |
| 420 |
AT1G67040 |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-1.839 |
-0.485 |
| 421 |
AT2G05100 |
AT2G05100
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.1, photosystem II light harvesting complex gene 2.1 |
-1.836 |
no data |
| 422 |
AT2G16890 |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-1.831 |
-0.058 |
| 423 |
AT5G24770 |
AT5G24770
|
ATVSP2, VSP2, vegetative storage protein 2 |
-1.804 |
no data |
| 424 |
AT3G29010 |
AT3G29010
|
[Biotin/lipoate A/B protein ligase family] |
-1.800 |
0.055 |
| 425 |
AT5G01900 |
AT5G01900
|
ATWRKY62, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 62, WRKY62, WRKY DNA-binding protein 62 |
-1.788 |
no data |
| 426 |
AT4G26200 |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-1.783 |
0.117 |
| 427 |
AT1G13270 |
AT1G13270
|
MAP1B, METHIONINE AMINOPEPTIDASE 1B, MAP1C, methionine aminopeptidase 1B |
-1.779 |
-0.026 |
| 428 |
AT4G10860 |
AT4G10860
|
unknown |
-1.779 |
no data |
| 429 |
AT1G67270 |
AT1G67270
|
[Zinc-finger domain of monoamine-oxidase A repressor R1 protein] |
-1.779 |
0.011 |
| 430 |
AT2G03540 |
AT2G03540
|
unknown |
-1.773 |
0.259 |
| 431 |
AT5G63290 |
AT5G63290
|
[Radical SAM superfamily protein] |
-1.773 |
0.002 |
| 432 |
AT1G80150 |
AT1G80150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.746 |
no data |
| 433 |
AT5G40000 |
AT5G40000
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.743 |
no data |
| 434 |
AT5G64120 |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-1.740 |
0.444 |
| 435 |
AT2G16060 |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-1.738 |
0.076 |
| 436 |
AT2G32630 |
AT2G32630
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-1.738 |
-0.096 |
| 437 |
AT5G39580 |
AT5G39580
|
[Peroxidase superfamily protein] |
-1.735 |
0.385 |
| 438 |
AT5G18840 |
AT5G18840
|
[Major facilitator superfamily protein] |
-1.729 |
-0.053 |
| 439 |
AT1G02390 |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-1.721 |
0.241 |
| 440 |
AT3G20360 |
AT3G20360
|
[TRAF-like family protein] |
-1.713 |
no data |
| 441 |
AT4G01630 |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-1.709 |
-0.027 |
| 442 |
AT2G47680 |
AT2G47680
|
[zinc finger (CCCH type) helicase family protein] |
-1.706 |
0.097 |
| 443 |
AT4G22470 |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-1.695 |
1.140 |
| 444 |
AT5G14920 |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-1.694 |
-0.190 |
| 445 |
AT1G09680 |
AT1G09680
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.671 |
-0.006 |
| 446 |
AT1G26790 |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
-1.663 |
-0.559 |
| 447 |
AT5G20830 |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
-1.654 |
0.139 |
| 448 |
AT3G18890 |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-1.651 |
-0.221 |
| 449 |
AT4G24080 |
AT4G24080
|
ALL1, aldolase like |
-1.651 |
no data |
| 450 |
AT2G26695 |
AT2G26695
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-1.651 |
no data |
| 451 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-1.645 |
1.010 |
| 452 |
AT5G65140 |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-1.640 |
0.162 |
| 453 |
AT3G23170 |
AT3G23170
|
unknown |
-1.635 |
0.469 |
| 454 |
AT1G10070 |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-1.623 |
0.773 |
| 455 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
-1.616 |
0.866 |
| 456 |
AT4G24040 |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-1.610 |
0.328 |
| 457 |
AT3G11210 |
AT3G11210
|
[SGNH hydrolase-type esterase superfamily protein] |
-1.605 |
0.057 |
| 458 |
AT5G09720 |
AT5G09720
|
[Magnesium transporter CorA-like family protein] |
-1.603 |
no data |
| 459 |
AT4G26050 |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-1.597 |
0.032 |
| 460 |
AT3G23250 |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
-1.569 |
1.110 |
| 461 |
AT2G26070 |
AT2G26070
|
AtRTE1, RTE1, REVERSION-TO-ETHYLENE SENSITIVITY1 |
-1.566 |
-0.040 |
| 462 |
AT5G63350 |
AT5G63350
|
unknown |
-1.564 |
no data |
| 463 |
AT4G18170 |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
-1.564 |
1.135 |
| 464 |
AT5G46680 |
AT5G46680
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-1.561 |
-0.004 |
| 465 |
AT4G10280 |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
-1.543 |
0.105 |
| 466 |
AT5G25260 |
AT5G25260
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-1.540 |
no data |
| 467 |
AT5G64550 |
AT5G64550
|
[loricrin-related] |
-1.537 |
no data |
| 468 |
AT4G18490 |
AT4G18490
|
unknown |
-1.527 |
-0.036 |
| 469 |
AT3G02840 |
AT3G02840
|
[ARM repeat superfamily protein] |
-1.522 |
0.864 |
| 470 |
AT2G26790 |
AT2G26790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.522 |
-0.042 |
| 471 |
AT1G17285 |
AT1G17285
|
unknown |
-1.521 |
no data |
| 472 |
AT4G04710 |
AT4G04710
|
CPK22, calcium-dependent protein kinase 22 |
-1.519 |
no data |
| 473 |
AT5G13910 |
AT5G13910
|
LEP, LEAFY PETIOLE |
-1.511 |
-0.006 |
| 474 |
ATCG00350 |
ATCG00350
|
PSAA |
-1.511 |
-0.070 |
| 475 |
AT2G18120 |
AT2G18120
|
SRS4, SHI-related sequence 4 |
-1.506 |
-0.216 |
| 476 |
AT1G56430 |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-1.501 |
-0.524 |
| 477 |
AT3G44880 |
AT3G44880
|
ACD1, ACCELERATED CELL DEATH 1, LLS1, LETHAL LEAF-SPOT 1 HOMOLOG, PAO, PHEOPHORBIDE A OXYGENASE |
-1.501 |
0.115 |
| 478 |
AT1G24420 |
AT1G24420
|
[HXXXD-type acyl-transferase family protein] |
-1.500 |
0.031 |
| 479 |
AT5G25610 |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-1.497 |
0.014 |
| 480 |
AT3G56430 |
AT3G56430
|
unknown |
-1.492 |
-0.035 |
| 481 |
AT1G60630 |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
-1.490 |
-0.121 |
| 482 |
AT1G72940 |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-1.490 |
0.139 |
| 483 |
AT1G71230 |
AT1G71230
|
AJH2, CSN5, CSN5B, COP9-signalosome 5B |
-1.487 |
0.009 |
| 484 |
AT4G18280 |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-1.482 |
0.239 |
| 485 |
AT5G09950 |
AT5G09950
|
MEF7, mitochondrial editing factor 7 |
-1.481 |
-0.027 |
| 486 |
AT1G29670 |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.480 |
0.075 |
| 487 |
AT1G17380 |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
-1.476 |
0.981 |
| 488 |
AT5G01542 |
AT5G01542
|
|
-1.476 |
no data |
| 489 |
AT1G67740 |
AT1G67740
|
PSBY, photosystem II BY, YCF32 |
-1.467 |
-0.062 |
| 490 |
AT3G18680 |
AT3G18680
|
[Amino acid kinase family protein] |
-1.467 |
-0.067 |
| 491 |
AT2G01340 |
AT2G01340
|
At17.1 |
-1.459 |
no data |
| 492 |
AT3G02100 |
AT3G02100
|
[UDP-Glycosyltransferase superfamily protein] |
-1.457 |
-0.022 |
| 493 |
AT5G35735 |
AT5G35735
|
[Auxin-responsive family protein] |
-1.457 |
0.634 |
| 494 |
AT2G30020 |
AT2G30020
|
[Protein phosphatase 2C family protein] |
-1.453 |
no data |
| 495 |
AT5G43120 |
AT5G43120
|
[ARM-repeat/Tetratricopeptide repeat (TPR)-like protein] |
-1.451 |
0.008 |
| 496 |
AT1G64430 |
AT1G64430
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.451 |
-0.188 |
| 497 |
AT1G77360 |
AT1G77360
|
APPR6, Arabidopsis pentatricopeptide repeat 6 |
-1.445 |
0.322 |
| 498 |
AT1G55670 |
AT1G55670
|
PSAG, photosystem I subunit G |
-1.445 |
0.002 |
| 499 |
AT1G52790 |
AT1G52790
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.438 |
0.040 |
| 500 |
AT3G30775 |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-1.433 |
-0.070 |
| 501 |
AT1G02180 |
AT1G02180
|
[ferredoxin-related] |
-1.429 |
-0.099 |
| 502 |
AT4G40020 |
AT4G40020
|
[Myosin heavy chain-related protein] |
-1.428 |
0.090 |
| 503 |
AT1G19720 |
AT1G19720
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-1.427 |
no data |
| 504 |
AT4G34138 |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
-1.423 |
0.245 |
| 505 |
AT5G50260 |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
-1.422 |
0.048 |
| 506 |
AT2G34390 |
AT2G34390
|
NIP2;1, NOD26-like intrinsic protein 2;1, NLM4, NOD26-LIKE MIP 4 |
-1.407 |
no data |
| 507 |
AT1G12400 |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-1.401 |
0.005 |
| 508 |
AT5G41290 |
AT5G41290
|
[Receptor-like protein kinase-related family protein] |
-1.397 |
no data |
| 509 |
AT1G19530 |
AT1G19530
|
unknown |
-1.395 |
0.053 |
| 510 |
AT5G39890 |
AT5G39890
|
unknown |
-1.395 |
0.020 |
| 511 |
AT1G50750 |
AT1G50750
|
[Plant mobile domain protein family] |
-1.392 |
-0.003 |
| 512 |
AT3G59440 |
AT3G59440
|
[Calcium-binding EF-hand family protein] |
-1.384 |
0.007 |
| 513 |
AT3G52740 |
AT3G52740
|
unknown |
-1.382 |
-0.157 |
| 514 |
AT3G55660 |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
-1.372 |
-0.035 |
| 515 |
AT1G13520 |
AT1G13520
|
unknown |
-1.370 |
no data |
| 516 |
ATCG01100 |
ATCG01100
|
NDHA |
-1.370 |
-0.072 |
| 517 |
AT4G15370 |
AT4G15370
|
BARS1, baruol synthase 1, PEN2, PENTACYCLIC TRITERPENE SYNTHASE 2 |
-1.367 |
0.027 |
| 518 |
AT3G12955 |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
-1.361 |
-0.047 |
| 519 |
ATCG00065 |
ATCG00065
|
RPS12, RIBOSOMAL PROTEIN S12, RPS12A, ribosomal protein S12A |
-1.361 |
-0.007 |
| 520 |
AT4G08150 |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
-1.358 |
-0.019 |
| 521 |
AT2G19190 |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-1.357 |
0.677 |
| 522 |
AT5G40590 |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.348 |
0.056 |
| 523 |
AT5G01540 |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-1.347 |
0.487 |
| 524 |
AT3G61470 |
AT3G61470
|
LHCA2, photosystem I light harvesting complex gene 2 |
-1.332 |
no data |
| 525 |
AT5G48540 |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
-1.326 |
0.214 |
| 526 |
AT3G49630 |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.323 |
-0.012 |
| 527 |
AT2G28630 |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-1.321 |
0.265 |
| 528 |
AT1G26380 |
AT1G26380
|
[FAD-binding Berberine family protein] |
-1.313 |
0.786 |
| 529 |
AT4G30340 |
AT4G30340
|
ATDGK7, diacylglycerol kinase 7, DGK7, diacylglycerol kinase 7 |
-1.311 |
0.152 |
| 530 |
AT3G28580 |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.301 |
0.181 |
| 531 |
AT5G45670 |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.299 |
-0.270 |
| 532 |
AT1G04310 |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-1.295 |
-0.217 |
| 533 |
AT5G50280 |
AT5G50280
|
EMB1006, embryo defective 1006 |
-1.294 |
-0.253 |
| 534 |
AT4G27450 |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-1.294 |
0.244 |
| 535 |
AT4G28590 |
AT4G28590
|
AtECB1, ECB1, early chloroplast biogenesis 1, MRL7, Mesophyll-cell RNAi Library line 7, SVR4, SUPPRESSOR OF VARIEGATION 4 |
-1.293 |
0.018 |
| 536 |
AT1G67105 |
AT1G67105
|
other RNA |
-1.293 |
no data |
| 537 |
AT4G33110 |
AT4G33110
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.289 |
no data |
| 538 |
AT1G13770 |
AT1G13770
|
RUS3, ROOT UV-B SENSITIVE 3 |
-1.283 |
no data |
| 539 |
AT1G13670 |
AT1G13670
|
unknown |
-1.280 |
-0.005 |
| 540 |
AT1G62670 |
AT1G62670
|
RPF2, rna processing factor 2 |
-1.279 |
-0.042 |
| 541 |
AT5G67310 |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
-1.279 |
0.605 |
| 542 |
AT2G24850 |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-1.278 |
0.651 |
| 543 |
AT5G22570 |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-1.275 |
0.144 |
| 544 |
AT2G07677 |
AT2G07677
|
[auxin-responsive factor (ARF) -related, low similarity to auxin response factor 10 (Arabidopsis thaliana) GI:6165644] |
-1.272 |
no data |
| 545 |
AT5G50690 |
AT5G50690
|
AtHSD7, hydroxysteroid dehydrogenase 7, HSD7, hydroxysteroid dehydrogenase 7 |
-1.272 |
no data |
| 546 |
AT3G21560 |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-1.265 |
-0.317 |
| 547 |
AT2G31380 |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
-1.261 |
-0.284 |
| 548 |
AT5G49850 |
AT5G49850
|
[Mannose-binding lectin superfamily protein] |
-1.261 |
no data |
| 549 |
AT1G21130 |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-1.258 |
0.498 |
| 550 |
AT5G04770 |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
-1.257 |
-0.050 |
| 551 |
AT3G19970 |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
-1.256 |
0.294 |
| 552 |
AT5G62520 |
AT5G62520
|
SRO5, similar to RCD one 5 |
-1.255 |
0.976 |
| 553 |
AT5G14650 |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-1.252 |
-0.041 |
| 554 |
AT3G17040 |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
-1.252 |
-0.220 |
| 555 |
AT3G10720 |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-1.251 |
0.020 |
| 556 |
AT1G04180 |
AT1G04180
|
YUC9, YUCCA 9 |
-1.246 |
0.030 |
| 557 |
AT5G05900 |
AT5G05900
|
[UDP-Glycosyltransferase superfamily protein] |
-1.246 |
0.054 |
| 558 |
AT1G05562 |
AT1G05562
|
Potential natural antisense gene |
-1.233 |
no data |
| 559 |
AT2G19800 |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-1.233 |
0.238 |
| 560 |
AT5G43890 |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
-1.232 |
-0.015 |
| 561 |
AT4G23700 |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-1.228 |
0.609 |
| 562 |
AT5G54960 |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-1.227 |
0.304 |
| 563 |
AT5G51620 |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-1.226 |
0.178 |
| 564 |
AT2G19590 |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-1.225 |
0.048 |
| 565 |
AT5G54470 |
AT5G54470
|
BBX29, B-box domain protein 29 |
-1.224 |
0.416 |
| 566 |
AT1G26870 |
AT1G26870
|
ANAC009, Arabidopsis NAC domain containing protein 9, FEZ, FEZ |
-1.219 |
0.020 |
| 567 |
AT1G05560 |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-1.215 |
-0.142 |
| 568 |
AT4G18197 |
AT4G18197
|
ATPUP7, ARABIDOPSIS THALIANA PURINE PERMEASE 7, PEX17, PEROXIN 17, PUP7, purine permease 7 |
-1.211 |
no data |
| 569 |
AT1G52690 |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-1.207 |
0.456 |
| 570 |
AT1G69230 |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-1.205 |
-0.069 |
| 571 |
AT3G17830 |
AT3G17830
|
DJA4, DNA J protein A4 |
-1.202 |
-0.062 |
| 572 |
AT3G21720 |
AT3G21720
|
ICL, isocitrate lyase |
-1.201 |
-0.037 |
| 573 |
AT2G35730 |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
-1.193 |
0.573 |
| 574 |
AT4G00910 |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-1.191 |
-0.019 |
| 575 |
AT5G45380 |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-1.179 |
0.074 |
| 576 |
AT2G05530 |
AT2G05530
|
[Glycine-rich protein family] |
-1.171 |
0.043 |
| 577 |
AT4G02330 |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-1.166 |
0.413 |
| 578 |
AT1G56570 |
AT1G56570
|
PGN, PENTATRICOPEPTIDE REPEAT PROTEIN FOR GERMINATION ON NaCl |
-1.166 |
-0.050 |
| 579 |
AT3G06430 |
AT3G06430
|
AtPPR2, EMB2750, embryo defective 2750, PPR2, pentatricopeptide repeat 2 |
-1.164 |
no data |
| 580 |
AT5G28770 |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-1.163 |
-0.287 |
| 581 |
AT5G43620 |
AT5G43620
|
[Pre-mRNA cleavage complex II] |
-1.163 |
no data |
| 582 |
AT5G02550 |
AT5G02550
|
unknown |
-1.162 |
0.135 |
| 583 |
AT2G46400 |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-1.156 |
0.401 |
| 584 |
AT3G03270 |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-1.153 |
0.008 |
| 585 |
AT4G17490 |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-1.150 |
0.489 |
| 586 |
AT3G43955 |
AT3G43955
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-1.150 |
no data |
| 587 |
AT3G44260 |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-1.150 |
0.486 |
| 588 |
AT4G14060 |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.148 |
-0.022 |
| 589 |
AT2G41230 |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-1.147 |
-0.139 |
| 590 |
AT3G55980 |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-1.145 |
0.365 |
| 591 |
ATCG00500 |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-1.144 |
-0.109 |
| 592 |
AT4G33950 |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-1.139 |
-0.123 |
| 593 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-1.133 |
0.581 |
| 594 |
AT1G29195 |
AT1G29195
|
unknown |
-1.133 |
0.081 |
| 595 |
AT1G55020 |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-1.133 |
0.029 |
| 596 |
AT5G10700 |
AT5G10700
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
-1.132 |
-0.018 |
| 597 |
AT2G44370 |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.131 |
0.514 |
| 598 |
AT3G03180 |
AT3G03180
|
[Got1/Sft2-like vescicle transport protein family] |
-1.128 |
0.030 |
| 599 |
AT4G32480 |
AT4G32480
|
unknown |
-1.127 |
no data |