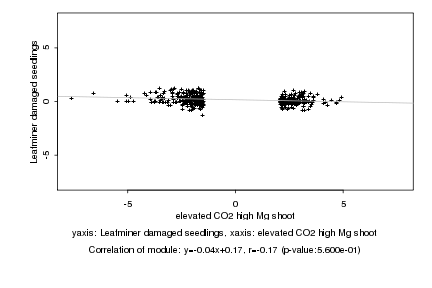
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
elevated CO2 high Mg shoot |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
AT3G57240 |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
4.899 |
0.378 |
| 2 |
AT5G09530 |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
4.791 |
0.178 |
| 3 |
AT1G50040 |
AT1G50040
|
unknown |
4.680 |
-0.112 |
| 4 |
AT5G61590 |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
4.657 |
-0.080 |
| 5 |
AT3G49110 |
AT3G49110
|
ATPCA, PEROXIDASE CA, ATPRX33, PRX33, PEROXIDASE 33, PRXCA, peroxidase CA |
4.444 |
no data |
| 6 |
AT2G38530 |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
4.414 |
0.188 |
| 7 |
AT2G32100 |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
4.240 |
-0.342 |
| 8 |
AT2G26400 |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
4.172 |
-0.002 |
| 9 |
AT3G01080 |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
4.049 |
0.225 |
| 10 |
AT4G34250 |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
4.043 |
-0.103 |
| 11 |
AT1G49130 |
AT1G49130
|
BBX17, B-box domain protein 17 |
3.911 |
no data |
| 12 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
3.800 |
0.726 |
| 13 |
AT1G69140 |
AT1G69140
|
[pseudogene] |
3.771 |
no data |
| 14 |
AT2G43580 |
AT2G43580
|
[Chitinase family protein] |
3.594 |
0.497 |
| 15 |
AT3G11340 |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
3.582 |
0.452 |
| 16 |
AT5G22570 |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
3.579 |
0.144 |
| 17 |
AT1G58225 |
AT1G58225
|
unknown |
3.561 |
no data |
| 18 |
AT5G64710 |
AT5G64710
|
[Putative endonuclease or glycosyl hydrolase] |
3.556 |
-0.012 |
| 19 |
AT1G19340 |
AT1G19340
|
[Methyltransferase MT-A70 family protein] |
3.516 |
-0.223 |
| 20 |
AT5G03350 |
AT5G03350
|
[Legume lectin family protein] |
3.510 |
-0.003 |
| 21 |
AT1G68520 |
AT1G68520
|
BBX14, B-box domain protein 14 |
3.480 |
-0.459 |
| 22 |
AT4G16260 |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
3.478 |
0.813 |
| 23 |
AT4G33610 |
AT4G33610
|
[glycine-rich protein] |
3.435 |
0.009 |
| 24 |
AT1G29440 |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
3.368 |
-0.720 |
| 25 |
AT1G78120 |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
3.357 |
-0.050 |
| 26 |
AT5G41040 |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
3.352 |
0.496 |
| 27 |
AT4G31330 |
AT4G31330
|
unknown |
3.348 |
-0.018 |
| 28 |
AT4G16700 |
AT4G16700
|
PSD1, phosphatidylserine decarboxylase 1 |
3.348 |
-0.084 |
| 29 |
AT1G21110 |
AT1G21110
|
IGMT3, indole glucosinolate O-methyltransferase 3 |
3.303 |
no data |
| 30 |
AT4G39580 |
AT4G39580
|
[Galactose oxidase/kelch repeat superfamily protein] |
3.303 |
no data |
| 31 |
AT2G25900 |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
3.298 |
0.066 |
| 32 |
AT2G33060 |
AT2G33060
|
AtRLP27, receptor like protein 27, RLP27, receptor like protein 27 |
3.296 |
no data |
| 33 |
AT2G21630 |
AT2G21630
|
[Sec23/Sec24 protein transport family protein] |
3.268 |
-0.007 |
| 34 |
AT4G16240 |
AT4G16240
|
unknown |
3.256 |
0.099 |
| 35 |
AT1G13609 |
AT1G13609
|
[Defensin-like (DEFL) family protein] |
3.238 |
no data |
| 36 |
AT3G22235 |
AT3G22235
|
unknown |
3.229 |
no data |
| 37 |
AT1G80440 |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
3.222 |
0.073 |
| 38 |
AT4G24000 |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
3.198 |
0.964 |
| 39 |
AT2G44380 |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
3.198 |
0.229 |
| 40 |
AT2G02950 |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
3.171 |
-0.783 |
| 41 |
AT3G55690 |
AT3G55690
|
unknown |
3.163 |
-0.130 |
| 42 |
AT1G26380 |
AT1G26380
|
[FAD-binding Berberine family protein] |
3.158 |
0.786 |
| 43 |
AT1G72230 |
AT1G72230
|
[Cupredoxin superfamily protein] |
3.157 |
-0.455 |
| 44 |
AT4G31441 |
AT4G31441
|
unknown |
3.157 |
no data |
| 45 |
AT1G06160 |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
3.157 |
0.524 |
| 46 |
AT1G74240 |
AT1G74240
|
[Mitochondrial substrate carrier family protein] |
3.157 |
-0.061 |
| 47 |
AT1G22330 |
AT1G22330
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
3.155 |
-0.472 |
| 48 |
AT3G22620 |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
3.105 |
0.780 |
| 49 |
AT3G23610 |
AT3G23610
|
DSPTP1, dual specificity protein phosphatase 1 |
3.105 |
-0.008 |
| 50 |
AT2G03670 |
AT2G03670
|
CDC48B, cell division cycle 48B |
3.105 |
-0.035 |
| 51 |
AT5G60022 |
AT5G60022
|
other RNA |
3.105 |
no data |
| 52 |
AT1G79310 |
AT1G79310
|
AtMC7, metacaspase 7, AtMCP2a, metacaspase 2a, MC7, metacaspase 7, MCP2a, metacaspase 2a |
3.105 |
0.463 |
| 53 |
AT3G53150 |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
3.105 |
0.182 |
| 54 |
AT1G15670 |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
3.105 |
0.435 |
| 55 |
AT3G58120 |
AT3G58120
|
ATBZIP61, BZIP61 |
3.101 |
-0.767 |
| 56 |
AT4G16000 |
AT4G16000
|
unknown |
3.091 |
0.661 |
| 57 |
AT1G14880 |
AT1G14880
|
AtPCR1, PCR1, PLANT CADMIUM RESISTANCE 1 |
3.086 |
no data |
| 58 |
AT2G18660 |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
3.079 |
0.162 |
| 59 |
AT3G48850 |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
3.051 |
0.586 |
| 60 |
AT4G10290 |
AT4G10290
|
[RmlC-like cupins superfamily protein] |
3.051 |
no data |
| 61 |
AT5G24080 |
AT5G24080
|
[Protein kinase superfamily protein] |
3.051 |
0.910 |
| 62 |
AT1G11320 |
AT1G11320
|
unknown |
3.017 |
-0.039 |
| 63 |
AT1G14640 |
AT1G14640
|
[SWAP (Suppressor-of-White-APricot)/surp domain-containing protein] |
3.016 |
0.111 |
| 64 |
AT1G79460 |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
3.013 |
-0.018 |
| 65 |
AT4G38080 |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
2.998 |
-0.040 |
| 66 |
AT3G47675 |
AT3G47675
|
unknown |
2.995 |
no data |
| 67 |
AT5G59662 |
AT5G59662
|
other RNA |
2.995 |
no data |
| 68 |
AT3G22600 |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
2.977 |
0.651 |
| 69 |
AT3G26790 |
AT3G26790
|
FUS3, FUSCA 3 |
2.977 |
-0.035 |
| 70 |
AT1G34340 |
AT1G34340
|
[alpha/beta-Hydrolases superfamily protein] |
2.972 |
0.144 |
| 71 |
AT3G26830 |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
2.966 |
0.856 |
| 72 |
AT3G50470 |
AT3G50470
|
HR3, homolog of RPW8 3, MLA10, INTRACELLULAR MILDEW A 10 |
2.956 |
no data |
| 73 |
AT3G22231 |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
2.940 |
-0.138 |
| 74 |
AT5G05840 |
AT5G05840
|
unknown |
2.939 |
0.052 |
| 75 |
AT1G67810 |
AT1G67810
|
SUFE2, sulfur E2 |
2.938 |
0.595 |
| 76 |
AT5G35777 |
AT5G35777
|
[copia-like retrotransposon family, has a 2.0e-67 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
2.937 |
no data |
| 77 |
AT1G65500 |
AT1G65500
|
unknown |
2.936 |
0.405 |
| 78 |
AT2G25200 |
AT2G25200
|
unknown |
2.919 |
0.051 |
| 79 |
AT5G26690 |
AT5G26690
|
[Heavy metal transport/detoxification superfamily protein ] |
2.918 |
no data |
| 80 |
AT2G23290 |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
2.913 |
-0.106 |
| 81 |
AT1G74940 |
AT1G74940
|
unknown |
2.900 |
-0.139 |
| 82 |
AT2G26440 |
AT2G26440
|
PME12, pectin methylesterase 12 |
2.886 |
0.509 |
| 83 |
AT4G02005 |
AT4G02005
|
other RNA |
2.877 |
no data |
| 84 |
AT5G23160 |
AT5G23160
|
unknown |
2.877 |
no data |
| 85 |
AT2G36050 |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
2.877 |
-0.044 |
| 86 |
AT4G38215 |
AT4G38215
|
other RNA |
2.877 |
no data |
| 87 |
AT5G49080 |
AT5G49080
|
EXT11, extensin 11 |
2.877 |
no data |
| 88 |
AT3G17180 |
AT3G17180
|
scpl33, serine carboxypeptidase-like 33 |
2.874 |
0.049 |
| 89 |
AT1G22360 |
AT1G22360
|
AtUGT85A2, UDP-glucosyl transferase 85A2, UGT85A2, UDP-glucosyl transferase 85A2 |
2.873 |
0.283 |
| 90 |
AT5G10310 |
AT5G10310
|
unknown |
2.852 |
-0.140 |
| 91 |
AT1G54010 |
AT1G54010
|
GLL23, GDSL-like lipase 23 |
2.849 |
no data |
| 92 |
AT1G36675 |
AT1G36675
|
[glycine-rich protein] |
2.840 |
no data |
| 93 |
AT1G19960 |
AT1G19960
|
unknown |
2.838 |
-0.237 |
| 94 |
AT3G15570 |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
2.822 |
-0.361 |
| 95 |
AT5G50260 |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
2.813 |
0.048 |
| 96 |
AT5G15430 |
AT5G15430
|
[Plant calmodulin-binding protein-related] |
2.813 |
no data |
| 97 |
AT3G51340 |
AT3G51340
|
[Eukaryotic aspartyl protease family protein] |
2.813 |
no data |
| 98 |
AT5G44920 |
AT5G44920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
2.808 |
0.006 |
| 99 |
AT1G15480 |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
2.806 |
-0.010 |
| 100 |
AT1G10690 |
AT1G10690
|
unknown |
2.803 |
0.055 |
| 101 |
AT1G26800 |
AT1G26800
|
[RING/U-box superfamily protein] |
2.786 |
-0.081 |
| 102 |
AT4G15233 |
AT4G15233
|
ABCG42, ATP-binding cassette G42 |
2.776 |
no data |
| 103 |
AT2G30770 |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
2.769 |
0.830 |
| 104 |
AT2G32830 |
AT2G32830
|
PHT1;5, phosphate transporter 1;5, PHT5, PHOSPHATE TRANSPORTER 5 |
2.763 |
0.151 |
| 105 |
AT3G08800 |
AT3G08800
|
SIEL, SHORT-ROOT interacting embryonic lethal |
2.763 |
-0.067 |
| 106 |
AT1G63160 |
AT1G63160
|
EMB2811, EMBRYO DEFECTIVE 2811, RFC2, replication factor C 2 |
2.750 |
0.055 |
| 107 |
AT3G16420 |
AT3G16420
|
JAL30, JACALIN-RELATED LECTIN 30, PBP1, PYK10-binding protein 1, PBPI |
2.750 |
no data |
| 108 |
AT5G09470 |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
2.749 |
0.089 |
| 109 |
AT4G14970 |
AT4G14970
|
unknown |
2.747 |
-0.048 |
| 110 |
AT2G33080 |
AT2G33080
|
AtRLP28, receptor like protein 28, RLP28, receptor like protein 28 |
2.747 |
0.138 |
| 111 |
AT1G02190 |
AT1G02190
|
[Fatty acid hydroxylase superfamily] |
2.736 |
-0.041 |
| 112 |
AT2G14560 |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
2.723 |
-0.129 |
| 113 |
AT3G04230 |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
2.716 |
0.147 |
| 114 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
2.716 |
0.597 |
| 115 |
AT1G36680 |
AT1G36680
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.6e-22 P-value blast match to At5g59620.1/14-257 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
2.716 |
no data |
| 116 |
AT1G65790 |
AT1G65790
|
ARK1, receptor kinase 1, RK1, receptor kinase 1 |
2.701 |
no data |
| 117 |
AT3G15680 |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
2.700 |
-0.235 |
| 118 |
AT1G43675 |
AT1G43675
|
[expressed protein] |
2.700 |
no data |
| 119 |
AT4G21903 |
AT4G21903
|
[MATE efflux family protein] |
2.689 |
no data |
| 120 |
AT5G61160 |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
2.683 |
1.075 |
| 121 |
AT5G62480 |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
2.678 |
0.287 |
| 122 |
AT4G30680 |
AT4G30680
|
[Initiation factor eIF-4 gamma, MA3] |
2.678 |
0.003 |
| 123 |
AT1G22500 |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
2.678 |
0.165 |
| 124 |
AT5G25570 |
AT5G25570
|
unknown |
2.678 |
0.001 |
| 125 |
AT1G73830 |
AT1G73830
|
BEE3, BR enhanced expression 3 |
2.678 |
-0.610 |
| 126 |
AT3G29250 |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
2.678 |
0.171 |
| 127 |
AT1G66960 |
AT1G66960
|
LUP5 |
2.673 |
0.037 |
| 128 |
AT2G16050 |
AT2G16050
|
[Cysteine/Histidine-rich C1 domain family protein] |
2.668 |
no data |
| 129 |
AT2G28120 |
AT2G28120
|
[Major facilitator superfamily protein] |
2.662 |
0.242 |
| 130 |
AT5G66480 |
AT5G66480
|
unknown |
2.658 |
no data |
| 131 |
AT5G40780 |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
2.656 |
0.436 |
| 132 |
AT1G77960 |
AT1G77960
|
unknown |
2.652 |
no data |
| 133 |
AT1G10657 |
AT1G10657
|
unknown |
2.638 |
no data |
| 134 |
AT4G37240 |
AT4G37240
|
unknown |
2.628 |
-0.277 |
| 135 |
AT1G80325 |
AT1G80325
|
other RNA |
2.618 |
no data |
| 136 |
AT4G23130 |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
2.603 |
0.060 |
| 137 |
AT3G19680 |
AT3G19680
|
unknown |
2.599 |
-0.221 |
| 138 |
AT2G25470 |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
2.595 |
0.467 |
| 139 |
AT5G54980 |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
2.585 |
-0.190 |
| 140 |
AT1G12260 |
AT1G12260
|
ANAC007, NAC 007, EMB2749, EMBRYO DEFECTIVE 2749, NAC007, NAC 007, VND4, VASCULAR RELATED NAC-DOMAIN PROTEIN 4 |
2.566 |
-0.056 |
| 141 |
AT1G29460 |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
2.566 |
-0.498 |
| 142 |
AT2G43010 |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
2.566 |
0.025 |
| 143 |
AT2G19190 |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
2.565 |
0.677 |
| 144 |
AT2G35470 |
AT2G35470
|
unknown |
2.565 |
-0.161 |
| 145 |
AT1G51790 |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
2.558 |
0.351 |
| 146 |
AT2G35070 |
AT2G35070
|
unknown |
2.554 |
no data |
| 147 |
AT3G14470 |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
2.549 |
-0.055 |
| 148 |
AT1G73810 |
AT1G73810
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
2.545 |
no data |
| 149 |
AT1G15790 |
AT1G15790
|
unknown |
2.526 |
0.249 |
| 150 |
AT5G66610 |
AT5G66610
|
DAR7, DA1-related protein 7 |
2.514 |
-0.052 |
| 151 |
AT1G61460 |
AT1G61460
|
[S-locus protein kinase, putative] |
2.512 |
-0.019 |
| 152 |
AT1G80320 |
AT1G80320
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
2.512 |
0.013 |
| 153 |
AT1G12160 |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
2.512 |
0.056 |
| 154 |
AT1G19230 |
AT1G19230
|
[Riboflavin synthase-like superfamily protein] |
2.511 |
-0.012 |
| 155 |
AT1G03220 |
AT1G03220
|
[Eukaryotic aspartyl protease family protein] |
2.511 |
no data |
| 156 |
AT4G13494 |
AT4G13494
|
MIR863A, microRNA863A |
2.511 |
no data |
| 157 |
AT3G62270 |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
2.503 |
-0.028 |
| 158 |
AT3G26440 |
AT3G26440
|
unknown |
2.498 |
0.250 |
| 159 |
AT4G02100 |
AT4G02100
|
[Heat shock protein DnaJ with tetratricopeptide repeat] |
2.488 |
no data |
| 160 |
AT4G10500 |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
2.477 |
-0.016 |
| 161 |
AT2G26020 |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
2.474 |
0.565 |
| 162 |
AT2G43590 |
AT2G43590
|
[Chitinase family protein] |
2.472 |
0.393 |
| 163 |
AT5G60280 |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
2.469 |
0.080 |
| 164 |
AT4G26542 |
AT4G26542
|
|
2.464 |
no data |
| 165 |
AT4G26540 |
AT4G26540
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
2.463 |
-0.206 |
| 166 |
AT3G47510 |
AT3G47510
|
unknown |
2.457 |
0.198 |
| 167 |
AT5G59410 |
AT5G59410
|
[Rab5-interacting family protein] |
2.456 |
-0.015 |
| 168 |
AT4G07830 |
AT4G07830
|
[gypsy-like retrotransposon family, has a 6.3e-159 P-value blast match to GB:CAA73042 polyprotein (Gypsy_Ty3-element) (Ananas comosus)] |
2.456 |
no data |
| 169 |
AT2G14830 |
AT2G14830
|
[Regulator of Vps4 activity in the MVB pathway protein] |
2.456 |
-0.000 |
| 170 |
AT4G21380 |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
2.453 |
0.298 |
| 171 |
AT5G55460 |
AT5G55460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
2.449 |
no data |
| 172 |
AT2G23130 |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
2.438 |
-0.619 |
| 173 |
AT2G21850 |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
2.436 |
0.061 |
| 174 |
AT1G70290 |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
2.432 |
0.123 |
| 175 |
AT5G51910 |
AT5G51910
|
[TCP family transcription factor ] |
2.428 |
-0.239 |
| 176 |
AT4G04940 |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
2.425 |
-0.081 |
| 177 |
AT5G38250 |
AT5G38250
|
[Protein kinase family protein] |
2.423 |
no data |
| 178 |
AT2G14460 |
AT2G14460
|
unknown |
2.419 |
0.043 |
| 179 |
AT3G07860 |
AT3G07860
|
[Ubiquitin-like superfamily protein] |
2.417 |
-0.112 |
| 180 |
AT4G13990 |
AT4G13990
|
[Exostosin family protein] |
2.416 |
-0.136 |
| 181 |
AT2G30790 |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
2.415 |
-0.147 |
| 182 |
AT3G32980 |
AT3G32980
|
[Peroxidase superfamily protein] |
2.406 |
no data |
| 183 |
AT2G37590 |
AT2G37590
|
ATDOF2.4, DOF2.4, DNA binding with one finger 2.4 |
2.398 |
0.016 |
| 184 |
AT5G19025 |
AT5G19025
|
[Ribosomal protein L34e superfamily protein] |
2.398 |
no data |
| 185 |
AT1G09794 |
AT1G09794
|
[Cox19 family protein (CHCH motif)] |
2.398 |
no data |
| 186 |
AT5G55790 |
AT5G55790
|
unknown |
2.398 |
-0.024 |
| 187 |
AT2G07719 |
AT2G07719
|
[Putative membrane lipoprotein] |
2.398 |
no data |
| 188 |
AT3G44990 |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
2.397 |
-0.738 |
| 189 |
AT3G45260 |
AT3G45260
|
[C2H2-like zinc finger protein] |
2.394 |
-0.001 |
| 190 |
AT3G53320 |
AT3G53320
|
unknown |
2.392 |
-0.107 |
| 191 |
AT2G20250 |
AT2G20250
|
unknown |
2.389 |
-0.049 |
| 192 |
AT2G23770 |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
2.388 |
0.046 |
| 193 |
AT3G28510 |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
2.369 |
0.062 |
| 194 |
AT1G02610 |
AT1G02610
|
[RING/FYVE/PHD zinc finger superfamily protein] |
2.359 |
no data |
| 195 |
AT3G27580 |
AT3G27580
|
ATPK7, D6PKL3, D6 PROTEIN KINASE LIKE 3 |
2.356 |
-0.184 |
| 196 |
AT1G27285 |
AT1G27285
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
2.355 |
-0.056 |
| 197 |
AT1G04930 |
AT1G04930
|
[hydroxyproline-rich glycoprotein family protein] |
2.355 |
0.039 |
| 198 |
AT1G31173 |
AT1G31173
|
MIR167D, microRNA167D |
2.355 |
no data |
| 199 |
AT4G15760 |
AT4G15760
|
MO1, monooxygenase 1 |
2.351 |
no data |
| 200 |
AT4G40042 |
AT4G40042
|
[Microsomal signal peptidase 12 kDa subunit (SPC12)] |
2.343 |
no data |
| 201 |
AT1G33102 |
AT1G33102
|
unknown |
2.343 |
no data |
| 202 |
AT1G29715 |
AT1G29715
|
[pseudogene] |
2.343 |
no data |
| 203 |
AT1G54510 |
AT1G54510
|
ATNEK1, NIMA-related serine/threonine kinase 1, NEK1, NIMA-related serine/threonine kinase 1 |
2.343 |
-0.032 |
| 204 |
AT1G17990 |
AT1G17990
|
[FMN-linked oxidoreductases superfamily protein] |
2.337 |
no data |
| 205 |
AT2G19990 |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
2.337 |
0.111 |
| 206 |
AT4G30800 |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
2.337 |
-0.042 |
| 207 |
AT4G11290 |
AT4G11290
|
[Peroxidase superfamily protein] |
2.335 |
-0.010 |
| 208 |
AT4G38210 |
AT4G38210
|
ATEXP20, ATEXPA20, expansin A20, ATHEXP ALPHA 1.23, EXP20, EXPANSIN 20, EXPA20, expansin A20 |
2.324 |
-0.051 |
| 209 |
AT4G21070 |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
2.324 |
-0.073 |
| 210 |
AT1G02230 |
AT1G02230
|
ANAC004, NAC domain containing protein 4, NAC004, NAC domain containing protein 4 |
2.318 |
no data |
| 211 |
AT3G49790 |
AT3G49790
|
[Carbohydrate-binding protein] |
2.316 |
0.154 |
| 212 |
AT2G24762 |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
2.314 |
-0.503 |
| 213 |
AT1G64600 |
AT1G64600
|
[methyltransferases] |
2.311 |
-0.074 |
| 214 |
AT4G27010 |
AT4G27010
|
EMB2788, EMBRYO DEFECTIVE 2788 |
2.298 |
-0.062 |
| 215 |
AT1G19330 |
AT1G19330
|
unknown |
2.297 |
-0.114 |
| 216 |
AT2G18700 |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
2.290 |
0.496 |
| 217 |
AT5G44430 |
AT5G44430
|
PDF1.2c, plant defensin 1.2C |
2.287 |
no data |
| 218 |
AT2G16660 |
AT2G16660
|
[Major facilitator superfamily protein] |
2.283 |
0.189 |
| 219 |
AT3G12500 |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
2.281 |
0.687 |
| 220 |
AT3G05250 |
AT3G05250
|
[RING/U-box superfamily protein] |
2.278 |
0.091 |
| 221 |
AT5G14580 |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
2.277 |
-0.116 |
| 222 |
AT3G55240 |
AT3G55240
|
unknown |
2.276 |
no data |
| 223 |
AT2G47130 |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
2.263 |
0.255 |
| 224 |
AT1G15830 |
AT1G15830
|
unknown |
2.262 |
-0.114 |
| 225 |
AT3G11760 |
AT3G11760
|
unknown |
2.262 |
-0.289 |
| 226 |
AT1G18020 |
AT1G18020
|
[FMN-linked oxidoreductases superfamily protein] |
2.256 |
no data |
| 227 |
AT2G44130 |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
2.254 |
0.069 |
| 228 |
AT3G16520 |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
2.250 |
-0.086 |
| 229 |
AT5G19190 |
AT5G19190
|
unknown |
2.246 |
no data |
| 230 |
AT5G05340 |
AT5G05340
|
PRX52, peroxidase 52 |
2.244 |
1.022 |
| 231 |
AT4G22490 |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
2.240 |
-0.003 |
| 232 |
AT5G64520 |
AT5G64520
|
ATXRCC2, XRCC2, homolog of X-ray repair cross complementing 2 |
2.239 |
no data |
| 233 |
AT3G13610 |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
2.237 |
0.215 |
| 234 |
AT3G63210 |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
2.230 |
-0.050 |
| 235 |
AT3G26500 |
AT3G26500
|
PIRL2, plant intracellular ras group-related LRR 2 |
2.229 |
no data |
| 236 |
AT2G39675 |
AT2G39675
|
TAS1C, trans-acting siRNA1C |
2.227 |
no data |
| 237 |
AT2G26790 |
AT2G26790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
2.225 |
-0.042 |
| 238 |
AT1G68190 |
AT1G68190
|
BBX27, B-box domain protein 27 |
2.225 |
-0.192 |
| 239 |
AT2G20670 |
AT2G20670
|
unknown |
2.212 |
-0.242 |
| 240 |
AT4G13495 |
AT4G13495
|
other RNA |
2.210 |
no data |
| 241 |
AT1G23390 |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
2.207 |
0.013 |
| 242 |
AT3G57700 |
AT3G57700
|
[Protein kinase superfamily protein] |
2.196 |
0.445 |
| 243 |
AT2G24750 |
AT2G24750
|
[pseudogene] |
2.196 |
no data |
| 244 |
AT3G06530 |
AT3G06530
|
[ARM repeat superfamily protein] |
2.196 |
-0.162 |
| 245 |
AT4G02660 |
AT4G02660
|
[Beige/BEACH domain ] |
2.187 |
-0.018 |
| 246 |
AT4G04510 |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
2.178 |
0.039 |
| 247 |
AT1G68780 |
AT1G68780
|
[RNI-like superfamily protein] |
2.178 |
-0.627 |
| 248 |
AT2G30930 |
AT2G30930
|
unknown |
2.176 |
0.235 |
| 249 |
AT3G57460 |
AT3G57460
|
[catalytics] |
2.176 |
0.679 |
| 250 |
AT2G36330 |
AT2G36330
|
[Uncharacterised protein family (UPF0497)] |
2.175 |
0.065 |
| 251 |
AT1G51850 |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
2.174 |
0.173 |
| 252 |
AT5G51440 |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
2.174 |
-0.687 |
| 253 |
AT4G13710 |
AT4G13710
|
[Pectin lyase-like superfamily protein] |
2.174 |
-0.120 |
| 254 |
AT5G37440 |
AT5G37440
|
[Chaperone DnaJ-domain superfamily protein] |
2.174 |
no data |
| 255 |
AT5G51170 |
AT5G51170
|
unknown |
2.174 |
0.024 |
| 256 |
AT2G17230 |
AT2G17230
|
EXL5, EXORDIUM like 5 |
2.168 |
-0.284 |
| 257 |
AT3G18610 |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
2.166 |
0.087 |
| 258 |
AT1G01070 |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
2.166 |
-0.044 |
| 259 |
AT3G18980 |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
2.164 |
-0.085 |
| 260 |
AT5G52740 |
AT5G52740
|
[Copper transport protein family] |
2.159 |
-0.133 |
| 261 |
AT1G13650 |
AT1G13650
|
unknown |
2.159 |
-0.608 |
| 262 |
AT4G23160 |
AT4G23160
|
CRK8, cysteine-rich RLK (RECEPTOR-like protein kinase) 8 |
2.159 |
no data |
| 263 |
AT2G34430 |
AT2G34430
|
LHB1B1, light-harvesting chlorophyll-protein complex II subunit B1, LHCB1.4, LIGHT-HARVESTING CHLOROPHYLL-PROTEIN COMPLEX II SUBUNIT B1 |
2.155 |
no data |
| 264 |
AT1G03300 |
AT1G03300
|
unknown |
2.154 |
-0.310 |
| 265 |
AT1G02920 |
AT1G02920
|
ATGST11, ARABIDOPSIS GLUTATHIONE S-TRANSFERASE 11, ATGSTF7, ATGSTF8, GST11, GLUTATHIONE S-TRANSFERASE 11, GSTF7, glutathione S-transferase 7 |
2.154 |
no data |
| 266 |
AT1G09080 |
AT1G09080
|
BIP3, binding protein 3 |
2.153 |
0.017 |
| 267 |
AT1G61370 |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
2.153 |
0.389 |
| 268 |
AT5G01230 |
AT5G01230
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.152 |
-0.078 |
| 269 |
AT5G56960 |
AT5G56960
|
[basic helix-loop-helix (bHLH) DNA-binding family protein] |
2.151 |
no data |
| 270 |
AT2G43000 |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
2.148 |
0.329 |
| 271 |
AT3G16530 |
AT3G16530
|
[Legume lectin family protein] |
2.144 |
0.406 |
| 272 |
AT2G40095 |
AT2G40095
|
[Alpha/beta hydrolase related protein] |
2.138 |
no data |
| 273 |
AT3G09490 |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
2.133 |
0.477 |
| 274 |
AT4G14550 |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
2.128 |
0.113 |
| 275 |
AT1G02300 |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
2.125 |
0.091 |
| 276 |
AT4G02540 |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
2.124 |
-0.155 |
| 277 |
AT4G18340 |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
2.120 |
0.237 |
| 278 |
AT5G34800 |
AT5G34800
|
[pseudogene] |
2.120 |
-0.086 |
| 279 |
AT3G53690 |
AT3G53690
|
[RING/U-box superfamily protein] |
2.120 |
0.058 |
| 280 |
AT5G51560 |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
2.120 |
-0.548 |
| 281 |
AT3G12502 |
AT3G12502
|
other RNA |
2.119 |
no data |
| 282 |
AT4G13570 |
AT4G13570
|
HTA4, histone H2A 4 |
2.117 |
-0.105 |
| 283 |
AT1G23800 |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
2.113 |
0.317 |
| 284 |
AT5G08330 |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
2.113 |
0.027 |
| 285 |
AT1G76960 |
AT1G76960
|
unknown |
2.104 |
0.352 |
| 286 |
AT4G02460 |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
2.104 |
-0.228 |
| 287 |
AT4G18270 |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
2.104 |
0.355 |
| 288 |
AT2G01008 |
AT2G01008
|
unknown |
2.098 |
no data |
| 289 |
AT1G67620 |
AT1G67620
|
[Lojap-related protein] |
2.096 |
0.074 |
| 290 |
AT1G15125 |
AT1G15125
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.094 |
no data |
| 291 |
AT1G09530 |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
2.089 |
0.040 |
| 292 |
AT4G04410 |
AT4G04410
|
[copia-like retrotransposon family, has a 1.7e-176 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
2.082 |
0.060 |
| 293 |
AT1G67050 |
AT1G67050
|
unknown |
2.080 |
-0.245 |
| 294 |
AT1G51860 |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
2.080 |
0.266 |
| 295 |
AT1G12090 |
AT1G12090
|
ELP, extensin-like protein |
2.074 |
-0.088 |
| 296 |
AT3G23080 |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
2.073 |
-0.070 |
| 297 |
AT5G24160 |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
2.072 |
0.207 |
| 298 |
AT4G04490 |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
2.067 |
0.298 |
| 299 |
AT2G14610 |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
2.065 |
0.214 |
| 300 |
AT5G55450 |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
2.064 |
0.048 |
| 301 |
AT4G25490 |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-7.644 |
0.333 |
| 302 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-6.613 |
0.837 |
| 303 |
AT1G73510 |
AT1G73510
|
unknown |
-5.520 |
0.082 |
| 304 |
AT4G13390 |
AT4G13390
|
EXT12, extensin 12 |
-5.097 |
0.004 |
| 305 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-5.094 |
0.615 |
| 306 |
AT5G51990 |
AT5G51990
|
CBF4, C-repeat-binding factor 4, DREB1D, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1D |
-5.004 |
0.090 |
| 307 |
AT4G24110 |
AT4G24110
|
unknown |
-4.875 |
0.395 |
| 308 |
AT5G13655 |
AT5G13655
|
unknown |
-4.856 |
no data |
| 309 |
AT4G24580 |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
-4.755 |
0.020 |
| 310 |
AT5G65300 |
AT5G65300
|
unknown |
-4.245 |
0.785 |
| 311 |
AT1G06030 |
AT1G06030
|
[pfkB-like carbohydrate kinase family protein] |
-4.204 |
no data |
| 312 |
AT5G05410 |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-4.158 |
0.635 |
| 313 |
AT5G38700 |
AT5G38700
|
unknown |
-3.983 |
0.219 |
| 314 |
AT2G22760 |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-3.982 |
0.899 |
| 315 |
AT1G01310 |
AT1G01310
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-3.923 |
-0.036 |
| 316 |
AT1G19040 |
AT1G19040
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-3.897 |
-0.026 |
| 317 |
AT2G32210 |
AT2G32210
|
unknown |
-3.848 |
no data |
| 318 |
AT3G14190 |
AT3G14190
|
CMR1, COPPER MODIFIED RESISTANCE 1 |
-3.799 |
-0.054 |
| 319 |
AT1G01460 |
AT1G01460
|
ATPIPK11, PIPK11 |
-3.799 |
0.029 |
| 320 |
AT3G49230 |
AT3G49230
|
unknown |
-3.798 |
-0.018 |
| 321 |
AT4G01360 |
AT4G01360
|
BPS3, BYPASS 3 |
-3.777 |
no data |
| 322 |
AT1G18310 |
AT1G18310
|
[glycosyl hydrolase family 81 protein] |
-3.732 |
0.072 |
| 323 |
AT3G10185 |
AT3G10185
|
[Gibberellin-regulated family protein] |
-3.732 |
no data |
| 324 |
AT1G02400 |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-3.731 |
0.932 |
| 325 |
AT4G27360 |
AT4G27360
|
[Dynein light chain type 1 family protein] |
-3.728 |
no data |
| 326 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
-3.695 |
0.866 |
| 327 |
AT1G45140 |
AT1G45140
|
[non-LTR retrotransposon family (LINE), has a 1.4e-37 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-3.678 |
no data |
| 328 |
AT3G28340 |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-3.664 |
0.375 |
| 329 |
AT4G01975 |
AT4G01975
|
[pseudogene] |
-3.663 |
no data |
| 330 |
AT2G39725 |
AT2G39725
|
[LYR family of Fe/S cluster biogenesis protein] |
-3.595 |
0.415 |
| 331 |
AT4G01390 |
AT4G01390
|
[TRAF-like family protein] |
-3.570 |
-0.005 |
| 332 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
-3.549 |
1.263 |
| 333 |
AT1G12610 |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-3.503 |
0.601 |
| 334 |
AT3G01840 |
AT3G01840
|
LYK2, LysM-containing receptor-like kinase 2 |
-3.499 |
0.154 |
| 335 |
AT5G03890 |
AT5G03890
|
unknown |
-3.475 |
no data |
| 336 |
AT1G70640 |
AT1G70640
|
[octicosapeptide/Phox/Bem1p (PB1) domain-containing protein] |
-3.474 |
no data |
| 337 |
AT5G05220 |
AT5G05220
|
unknown |
-3.436 |
0.239 |
| 338 |
AT3G09620 |
AT3G09620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-3.431 |
-0.020 |
| 339 |
AT3G05800 |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-3.416 |
0.447 |
| 340 |
AT2G25460 |
AT2G25460
|
unknown |
-3.364 |
0.749 |
| 341 |
AT4G05048 |
AT4G05048
|
U49.1 |
-3.344 |
no data |
| 342 |
AT5G47130 |
AT5G47130
|
[Bax inhibitor-1 family protein] |
-3.344 |
-0.002 |
| 343 |
AT3G04010 |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
-3.337 |
0.967 |
| 344 |
AT3G52450 |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-3.311 |
0.373 |
| 345 |
AT2G47050 |
AT2G47050
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-3.253 |
-0.033 |
| 346 |
AT1G24580 |
AT1G24580
|
[RING/U-box superfamily protein] |
-3.180 |
no data |
| 347 |
AT2G30020 |
AT2G30020
|
[Protein phosphatase 2C family protein] |
-3.177 |
no data |
| 348 |
AT1G12570 |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-3.154 |
-0.043 |
| 349 |
AT3G28600 |
AT3G28600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-3.141 |
0.149 |
| 350 |
AT4G35655 |
AT4G35655
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-3.119 |
no data |
| 351 |
AT5G49330 |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-3.118 |
-0.297 |
| 352 |
AT3G48520 |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-3.058 |
1.091 |
| 353 |
AT5G63160 |
AT5G63160
|
BT1, BTB and TAZ domain protein 1 |
-3.028 |
no data |
| 354 |
AT1G18300 |
AT1G18300
|
atnudt4, nudix hydrolase homolog 4, NUDT4, nudix hydrolase homolog 4 |
-3.024 |
no data |
| 355 |
AT5G26790 |
AT5G26790
|
unknown |
-3.019 |
-0.295 |
| 356 |
AT1G61340 |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-2.983 |
1.153 |
| 357 |
AT1G65390 |
AT1G65390
|
ATPP2-A5, phloem protein 2 A5, PP2-A5, phloem protein 2 A5 |
-2.948 |
0.726 |
| 358 |
AT4G17490 |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-2.946 |
0.489 |
| 359 |
AT5G45630 |
AT5G45630
|
unknown |
-2.941 |
0.353 |
| 360 |
AT4G29780 |
AT4G29780
|
unknown |
-2.923 |
0.791 |
| 361 |
AT1G31840 |
AT1G31840
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.909 |
-0.021 |
| 362 |
AT2G36650 |
AT2G36650
|
unknown |
-2.906 |
0.252 |
| 363 |
AT1G05575 |
AT1G05575
|
unknown |
-2.896 |
1.162 |
| 364 |
AT2G17830 |
AT2G17830
|
[F-box and associated interaction domains-containing protein] |
-2.840 |
no data |
| 365 |
AT5G43650 |
AT5G43650
|
BHLH92 |
-2.832 |
no data |
| 366 |
AT1G52890 |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-2.830 |
1.262 |
| 367 |
AT3G16150 |
AT3G16150
|
ASPGB1, asparaginase B1 |
-2.828 |
-0.027 |
| 368 |
AT3G56090 |
AT3G56090
|
ATFER3, ferritin 3, FER3, ferritin 3 |
-2.778 |
-0.073 |
| 369 |
AT4G05049 |
AT4G05049
|
|
-2.776 |
no data |
| 370 |
AT4G13395 |
AT4G13395
|
DVL10, DEVIL 10, RTFL12, ROTUNDIFOLIA like 12 |
-2.741 |
no data |
| 371 |
AT3G04800 |
AT3G04800
|
ATTIM23-3, translocase inner membrane subunit 23-3, TIM23-3, translocase inner membrane subunit 23-3 |
-2.738 |
-0.031 |
| 372 |
AT1G30390 |
AT1G30390
|
[non-LTR retrotransposon family (LINE), has a 1.9e-17 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-2.730 |
no data |
| 373 |
AT4G08145 |
AT4G08145
|
unknown |
-2.717 |
no data |
| 374 |
AT3G49130 |
AT3G49130
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
-2.717 |
0.312 |
| 375 |
AT5G03210 |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
-2.712 |
0.760 |
| 376 |
AT4G35480 |
AT4G35480
|
RHA3B, RING-H2 finger A3B |
-2.692 |
0.718 |
| 377 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-2.691 |
0.507 |
| 378 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-2.644 |
0.511 |
| 379 |
AT3G02840 |
AT3G02840
|
[ARM repeat superfamily protein] |
-2.638 |
0.864 |
| 380 |
AT5G16360 |
AT5G16360
|
[NC domain-containing protein-related] |
-2.636 |
no data |
| 381 |
AT4G20390 |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
-2.632 |
0.027 |
| 382 |
AT2G01150 |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-2.620 |
0.365 |
| 383 |
AT5G03204 |
AT5G03204
|
unknown |
-2.608 |
no data |
| 384 |
AT3G09590 |
AT3G09590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-2.594 |
0.076 |
| 385 |
AT4G12410 |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-2.593 |
0.444 |
| 386 |
AT2G18193 |
AT2G18193
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-2.579 |
no data |
| 387 |
AT4G13420 |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-2.574 |
0.007 |
| 388 |
AT4G29850 |
AT4G29850
|
Eukaryotic protein of unknown function (DUF872) |
-2.527 |
no data |
| 389 |
AT4G28140 |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
-2.525 |
1.203 |
| 390 |
AT3G23170 |
AT3G23170
|
unknown |
-2.516 |
0.469 |
| 391 |
AT1G01520 |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-2.489 |
-0.678 |
| 392 |
AT3G46080 |
AT3G46080
|
[C2H2-type zinc finger family protein] |
-2.481 |
no data |
| 393 |
AT5G66080 |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-2.481 |
0.107 |
| 394 |
AT3G30460 |
AT3G30460
|
[RING/U-box superfamily protein] |
-2.475 |
-0.244 |
| 395 |
AT4G26150 |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-2.475 |
-0.319 |
| 396 |
AT1G19210 |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-2.459 |
0.304 |
| 397 |
AT5G40000 |
AT5G40000
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-2.456 |
no data |
| 398 |
AT5G41080 |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-2.454 |
0.013 |
| 399 |
AT1G27730 |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-2.453 |
0.833 |
| 400 |
AT1G74930 |
AT1G74930
|
ORA47 |
-2.444 |
0.467 |
| 401 |
AT1G63030 |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
-2.440 |
0.038 |
| 402 |
AT1G42980 |
AT1G42980
|
[Actin-binding FH2 (formin homology 2) family protein] |
-2.424 |
no data |
| 403 |
AT3G43250 |
AT3G43250
|
unknown |
-2.420 |
0.876 |
| 404 |
AT1G63180 |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-2.416 |
-0.244 |
| 405 |
AT1G23340 |
AT1G23340
|
unknown |
-2.407 |
-0.150 |
| 406 |
AT3G56790 |
AT3G56790
|
[RNA splicing factor-related] |
-2.404 |
no data |
| 407 |
AT4G14690 |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-2.393 |
-0.157 |
| 408 |
AT2G17660 |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-2.377 |
0.422 |
| 409 |
AT1G78390 |
AT1G78390
|
ATNCED9, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9, NCED9, nine-cis-epoxycarotenoid dioxygenase 9 |
-2.361 |
0.024 |
| 410 |
AT3G23630 |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
-2.358 |
-0.084 |
| 411 |
AT3G52480 |
AT3G52480
|
unknown |
-2.348 |
-0.112 |
| 412 |
AT1G19190 |
AT1G19190
|
[alpha/beta-Hydrolases superfamily protein] |
-2.343 |
0.148 |
| 413 |
AT1G76600 |
AT1G76600
|
unknown |
-2.322 |
0.435 |
| 414 |
AT1G05100 |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
-2.314 |
0.778 |
| 415 |
AT3G14440 |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-2.311 |
1.073 |
| 416 |
AT1G03180 |
AT1G03180
|
unknown |
-2.306 |
0.027 |
| 417 |
AT1G73540 |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-2.303 |
0.216 |
| 418 |
AT1G11670 |
AT1G11670
|
[MATE efflux family protein] |
-2.300 |
0.454 |
| 419 |
AT1G32920 |
AT1G32920
|
unknown |
-2.297 |
0.578 |
| 420 |
AT3G61190 |
AT3G61190
|
BAP1, BON association protein 1 |
-2.286 |
0.828 |
| 421 |
AT1G06180 |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-2.278 |
0.291 |
| 422 |
AT1G09250 |
AT1G09250
|
AIF4, ATBS1 Interacting Factor 4 |
-2.276 |
0.077 |
| 423 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
-2.257 |
no data |
| 424 |
AT5G45340 |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-2.257 |
0.606 |
| 425 |
AT5G11260 |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
-2.255 |
-0.425 |
| 426 |
AT1G75490 |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
-2.255 |
0.034 |
| 427 |
AT3G54510 |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-2.255 |
-0.004 |
| 428 |
AT1G07150 |
AT1G07150
|
MAPKKK13, mitogen-activated protein kinase kinase kinase 13 |
-2.250 |
0.024 |
| 429 |
AT3G24518 |
AT3G24518
|
other RNA |
-2.243 |
no data |
| 430 |
AT5G44110 |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-2.240 |
-0.188 |
| 431 |
AT3G24520 |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-2.238 |
0.467 |
| 432 |
AT4G34412 |
AT4G34412
|
unknown |
-2.227 |
no data |
| 433 |
AT2G22880 |
AT2G22880
|
[VQ motif-containing protein] |
-2.220 |
1.092 |
| 434 |
AT2G47770 |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-2.194 |
0.261 |
| 435 |
AT1G05136 |
AT1G05136
|
unknown |
-2.194 |
no data |
| 436 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
-2.177 |
0.866 |
| 437 |
AT5G20250 |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-2.173 |
-0.061 |
| 438 |
AT5G01595 |
AT5G01595
|
other RNA |
-2.165 |
no data |
| 439 |
AT5G62520 |
AT5G62520
|
SRO5, similar to RCD one 5 |
-2.161 |
0.976 |
| 440 |
AT5G42760 |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-2.158 |
-0.796 |
| 441 |
AT2G35930 |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-2.152 |
0.595 |
| 442 |
AT3G55980 |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-2.151 |
0.365 |
| 443 |
AT1G30190 |
AT1G30190
|
unknown |
-2.151 |
no data |
| 444 |
AT1G74450 |
AT1G74450
|
unknown |
-2.146 |
0.222 |
| 445 |
AT2G40000 |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-2.144 |
0.347 |
| 446 |
AT5G43620 |
AT5G43620
|
[Pre-mRNA cleavage complex II] |
-2.135 |
no data |
| 447 |
AT5G38070 |
AT5G38070
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-2.134 |
-0.076 |
| 448 |
AT1G33760 |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-2.132 |
0.238 |
| 449 |
AT1G05550 |
AT1G05550
|
unknown |
-2.130 |
-0.002 |
| 450 |
AT3G53790 |
AT3G53790
|
TRFL4, TRF-like 4 |
-2.126 |
0.080 |
| 451 |
AT2G35637 |
AT2G35637
|
other RNA |
-2.125 |
no data |
| 452 |
AT1G18400 |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-2.124 |
-0.227 |
| 453 |
AT5G36925 |
AT5G36925
|
unknown |
-2.120 |
no data |
| 454 |
AT4G30280 |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-2.117 |
0.697 |
| 455 |
AT5G06990 |
AT5G06990
|
unknown |
-2.095 |
no data |
| 456 |
AT1G22190 |
AT1G22190
|
RAP2.4, related to AP2 4 |
-2.086 |
0.245 |
| 457 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-2.082 |
1.010 |
| 458 |
AT4G17310 |
AT4G17310
|
unknown |
-2.080 |
0.116 |
| 459 |
AT1G64760 |
AT1G64760
|
[O-Glycosyl hydrolases family 17 protein] |
-2.077 |
no data |
| 460 |
AT1G74670 |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-2.075 |
-0.422 |
| 461 |
AT5G01600 |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-2.074 |
0.217 |
| 462 |
AT1G77020 |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
-2.074 |
-0.403 |
| 463 |
AT5G45440 |
AT5G45440
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-2.070 |
no data |
| 464 |
AT3G03776 |
AT3G03776
|
[hydroxyproline-rich glycoprotein family protein] |
-2.066 |
no data |
| 465 |
AT4G22790 |
AT4G22790
|
[MATE efflux family protein] |
-2.057 |
-0.034 |
| 466 |
AT5G17350 |
AT5G17350
|
unknown |
-2.055 |
0.193 |
| 467 |
AT5G18670 |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-2.055 |
-0.748 |
| 468 |
AT4G34550 |
AT4G34550
|
unknown |
-2.049 |
no data |
| 469 |
AT2G35640 |
AT2G35640
|
[Homeodomain-like superfamily protein] |
-2.038 |
no data |
| 470 |
AT1G16370 |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
-2.037 |
0.830 |
| 471 |
AT3G02140 |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-2.031 |
0.363 |
| 472 |
AT5G14760 |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-2.024 |
-0.666 |
| 473 |
AT2G14900 |
AT2G14900
|
[Gibberellin-regulated family protein] |
-2.021 |
-0.180 |
| 474 |
AT1G49450 |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-2.020 |
1.018 |
| 475 |
AT3G58070 |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-2.016 |
-0.537 |
| 476 |
AT5G17050 |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
-2.013 |
-0.258 |
| 477 |
AT5G21960 |
AT5G21960
|
[Integrase-type DNA-binding superfamily protein] |
-2.006 |
no data |
| 478 |
AT5G47060 |
AT5G47060
|
unknown |
-2.003 |
0.295 |
| 479 |
AT3G23250 |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
-2.001 |
1.110 |
| 480 |
AT4G30350 |
AT4G30350
|
SMXL2, SMAX1-like 2 |
-1.996 |
0.520 |
| 481 |
AT2G27080 |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-1.994 |
0.560 |
| 482 |
AT5G62360 |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.987 |
1.055 |
| 483 |
AT3G44260 |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-1.983 |
0.486 |
| 484 |
AT5G63130 |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
-1.978 |
0.413 |
| 485 |
AT3G25250 |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
-1.977 |
0.406 |
| 486 |
AT4G39670 |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
-1.962 |
0.939 |
| 487 |
AT4G11280 |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-1.959 |
0.310 |
| 488 |
AT2G26695 |
AT2G26695
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-1.956 |
no data |
| 489 |
AT4G36950 |
AT4G36950
|
MAPKKK21, mitogen-activated protein kinase kinase kinase 21 |
-1.953 |
no data |
| 490 |
AT4G24972 |
AT4G24972
|
TPD1, TAPETUM DETERMINANT 1 |
-1.948 |
no data |
| 491 |
AT3G27210 |
AT3G27210
|
unknown |
-1.944 |
0.793 |
| 492 |
AT5G54470 |
AT5G54470
|
BBX29, B-box domain protein 29 |
-1.943 |
0.416 |
| 493 |
AT3G62260 |
AT3G62260
|
[Protein phosphatase 2C family protein] |
-1.939 |
0.159 |
| 494 |
AT1G26790 |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
-1.928 |
-0.559 |
| 495 |
AT2G41640 |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-1.926 |
0.432 |
| 496 |
AT2G20880 |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-1.926 |
0.897 |
| 497 |
AT1G66160 |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-1.923 |
0.915 |
| 498 |
AT2G46940 |
AT2G46940
|
unknown |
-1.921 |
0.079 |
| 499 |
AT2G06045 |
AT2G06045
|
[copia-like retrotransposon family, has a 2.2e-12 P-value blast match to GB:AAC02672 polyprotein (Ty1_Copia-element) (Arabidopsis arenosa)] |
-1.908 |
no data |
| 500 |
AT3G25710 |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-1.893 |
-0.276 |
| 501 |
AT3G56290 |
AT3G56290
|
unknown |
-1.886 |
-0.250 |
| 502 |
AT5G24120 |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-1.880 |
-0.152 |
| 503 |
AT3G21560 |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-1.865 |
-0.317 |
| 504 |
AT1G23710 |
AT1G23710
|
unknown |
-1.858 |
0.294 |
| 505 |
AT4G27940 |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
-1.856 |
-0.161 |
| 506 |
AT2G38240 |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.855 |
0.843 |
| 507 |
AT1G01480 |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
-1.845 |
0.563 |
| 508 |
AT5G05180 |
AT5G05180
|
unknown |
-1.844 |
-0.140 |
| 509 |
AT5G24390 |
AT5G24390
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
-1.844 |
-0.058 |
| 510 |
AT3G15060 |
AT3G15060
|
AtRABA1g, RAB GTPase homolog A1G, RABA1g, RAB GTPase homolog A1G |
-1.840 |
0.146 |
| 511 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-1.833 |
0.581 |
| 512 |
AT1G11210 |
AT1G11210
|
unknown |
-1.833 |
0.762 |
| 513 |
AT3G24760 |
AT3G24760
|
[Galactose oxidase/kelch repeat superfamily protein] |
-1.826 |
-0.079 |
| 514 |
AT1G09950 |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
-1.820 |
0.481 |
| 515 |
AT1G67070 |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
-1.813 |
0.098 |
| 516 |
AT4G36010 |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-1.808 |
0.998 |
| 517 |
AT4G22758 |
AT4G22758
|
unknown |
-1.801 |
no data |
| 518 |
AT5G01380 |
AT5G01380
|
[Homeodomain-like superfamily protein] |
-1.796 |
0.213 |
| 519 |
AT4G01985 |
AT4G01985
|
unknown |
-1.791 |
no data |
| 520 |
AT1G17060 |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-1.789 |
0.164 |
| 521 |
AT1G36370 |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
-1.786 |
0.001 |
| 522 |
AT2G36220 |
AT2G36220
|
unknown |
-1.786 |
0.605 |
| 523 |
AT4G22620 |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
-1.784 |
0.453 |
| 524 |
AT4G23750 |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-1.777 |
-0.290 |
| 525 |
AT3G46620 |
AT3G46620
|
unknown |
-1.771 |
0.256 |
| 526 |
AT5G48250 |
AT5G48250
|
BBX8, B-box domain protein 8 |
-1.755 |
0.001 |
| 527 |
AT1G01720 |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
-1.753 |
0.401 |
| 528 |
AT2G21200 |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-1.753 |
-0.213 |
| 529 |
AT4G05100 |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-1.753 |
1.214 |
| 530 |
AT3G46660 |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
-1.752 |
1.298 |
| 531 |
AT3G46658 |
AT3G46658
|
other RNA |
-1.752 |
no data |
| 532 |
AT2G40140 |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-1.751 |
0.411 |
| 533 |
AT1G67470 |
AT1G67470
|
[Protein kinase superfamily protein] |
-1.740 |
0.066 |
| 534 |
AT3G48280 |
AT3G48280
|
CYP71A25, cytochrome P450, family 71, subfamily A, polypeptide 25 |
-1.732 |
no data |
| 535 |
AT1G04770 |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.716 |
-0.176 |
| 536 |
AT1G67856 |
AT1G67856
|
[RING/U-box superfamily protein] |
-1.715 |
no data |
| 537 |
AT1G67855 |
AT1G67855
|
unknown |
-1.714 |
no data |
| 538 |
AT3G24460 |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-1.706 |
-0.579 |
| 539 |
AT4G33440 |
AT4G33440
|
[Pectin lyase-like superfamily protein] |
-1.701 |
-0.338 |
| 540 |
AT5G04340 |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-1.697 |
0.847 |
| 541 |
AT5G61800 |
AT5G61800
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.697 |
-0.034 |
| 542 |
AT2G36630 |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
-1.697 |
-0.121 |
| 543 |
AT1G65450 |
AT1G65450
|
GLC, GLAUCE |
-1.696 |
0.006 |
| 544 |
AT5G64750 |
AT5G64750
|
ABR1, ABA REPRESSOR1 |
-1.694 |
no data |
| 545 |
AT1G21090 |
AT1G21090
|
[Cupredoxin superfamily protein] |
-1.693 |
-0.065 |
| 546 |
AT1G19180 |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-1.688 |
0.900 |
| 547 |
AT5G48850 |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-1.687 |
-0.023 |
| 548 |
AT5G11070 |
AT5G11070
|
unknown |
-1.686 |
-0.016 |
| 549 |
AT4G27350 |
AT4G27350
|
unknown |
-1.682 |
0.028 |
| 550 |
AT5G49480 |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
-1.678 |
0.174 |
| 551 |
AT4G17615 |
AT4G17615
|
ATCBL1, ARABIDOPSIS THALIANA CALCINEURIN B-LIKE PROTEIN, CBL1, calcineurin B-like protein 1, SCABP5, SOS3-LIKE CALCIUM BINDING PROTEIN 5 |
-1.675 |
0.485 |
| 552 |
AT4G22470 |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-1.675 |
1.140 |
| 553 |
AT1G18740 |
AT1G18740
|
unknown |
-1.669 |
0.238 |
| 554 |
AT2G18680 |
AT2G18680
|
unknown |
-1.665 |
no data |
| 555 |
AT4G27657 |
AT4G27657
|
unknown |
-1.660 |
0.474 |
| 556 |
AT2G45900 |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
-1.660 |
-0.271 |
| 557 |
AT1G10560 |
AT1G10560
|
ATPUB18, ARABIDOPSIS THALIANA PLANT U-BOX 18, PUB18, plant U-box 18 |
-1.658 |
0.359 |
| 558 |
AT5G59010 |
AT5G59010
|
BSK5, brassinosteroid-signaling kinase 5 |
-1.650 |
-0.101 |
| 559 |
AT1G62290 |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-1.650 |
-0.209 |
| 560 |
AT4G30610 |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-1.646 |
-0.707 |
| 561 |
AT1G78170 |
AT1G78170
|
unknown |
-1.637 |
0.087 |
| 562 |
AT1G22570 |
AT1G22570
|
[Major facilitator superfamily protein] |
-1.636 |
0.812 |
| 563 |
AT1G52565 |
AT1G52565
|
unknown |
-1.629 |
no data |
| 564 |
AT5G11090 |
AT5G11090
|
[serine-rich protein-related] |
-1.621 |
0.104 |
| 565 |
AT1G22690 |
AT1G22690
|
[Gibberellin-regulated family protein] |
-1.620 |
-0.254 |
| 566 |
AT2G25690 |
AT2G25690
|
unknown |
-1.612 |
0.123 |
| 567 |
AT3G17510 |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
-1.607 |
-0.005 |
| 568 |
AT1G50750 |
AT1G50750
|
[Plant mobile domain protein family] |
-1.604 |
-0.003 |
| 569 |
AT2G35950 |
AT2G35950
|
EDA12, embryo sac development arrest 12 |
-1.603 |
-0.040 |
| 570 |
AT1G64380 |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-1.602 |
1.093 |
| 571 |
AT3G19240 |
AT3G19240
|
[Vacuolar import/degradation, Vid27-related protein] |
-1.601 |
0.096 |
| 572 |
AT5G54130 |
AT5G54130
|
[Calcium-binding endonuclease/exonuclease/phosphatase family] |
-1.601 |
-0.425 |
| 573 |
AT1G15010 |
AT1G15010
|
unknown |
-1.599 |
0.917 |
| 574 |
AT3G56300 |
AT3G56300
|
[Cysteinyl-tRNA synthetase, class Ia family protein] |
-1.597 |
-0.151 |
| 575 |
AT4G16140 |
AT4G16140
|
[proline-rich family protein] |
-1.596 |
-0.205 |
| 576 |
AT5G64310 |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
-1.595 |
0.378 |
| 577 |
AT4G37610 |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-1.593 |
-0.348 |
| 578 |
AT3G63060 |
AT3G63060
|
EDL3, EID1-like 3 |
-1.593 |
0.742 |
| 579 |
AT1G21550 |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-1.588 |
0.642 |
| 580 |
AT2G27420 |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-1.571 |
-1.219 |
| 581 |
AT3G56880 |
AT3G56880
|
[VQ motif-containing protein] |
-1.569 |
0.384 |
| 582 |
AT1G72210 |
AT1G72210
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.563 |
no data |
| 583 |
AT2G28200 |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-1.562 |
-0.492 |
| 584 |
AT4G24390 |
AT4G24390
|
AFB4, auxin signaling F-box 4 |
-1.561 |
0.042 |
| 585 |
AT5G14105 |
AT5G14105
|
unknown |
-1.558 |
0.076 |
| 586 |
AT2G44080 |
AT2G44080
|
ARL, ARGOS-like |
-1.553 |
0.053 |
| 587 |
AT4G09970 |
AT4G09970
|
unknown |
-1.552 |
-0.301 |
| 588 |
AT1G66180 |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-1.549 |
-0.070 |
| 589 |
AT1G15700 |
AT1G15700
|
ATPC2 |
-1.547 |
-0.161 |
| 590 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-1.545 |
-0.039 |
| 591 |
AT3G57770 |
AT3G57770
|
[Protein kinase superfamily protein] |
-1.544 |
0.043 |
| 592 |
AT4G27652 |
AT4G27652
|
unknown |
-1.539 |
0.727 |
| 593 |
AT4G22780 |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-1.533 |
0.746 |
| 594 |
AT4G25700 |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
-1.527 |
-0.359 |
| 595 |
AT4G33040 |
AT4G33040
|
[Thioredoxin superfamily protein] |
-1.520 |
-0.234 |
| 596 |
AT2G44840 |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-1.513 |
1.058 |
| 597 |
AT5G62165 |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-1.512 |
0.227 |
| 598 |
AT3G04640 |
AT3G04640
|
[glycine-rich protein] |
-1.508 |
0.826 |
| 599 |
AT2G35290 |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-1.507 |
0.788 |
| 600 |
AT4G40020 |
AT4G40020
|
[Myosin heavy chain-related protein] |
-1.506 |
0.090 |