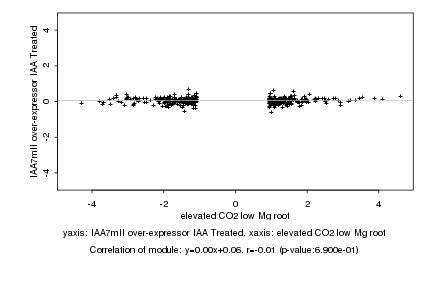
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
elevated CO2 low Mg root |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
AT2G29220 |
AT2G29220
|
LecRK-III.1, L-type lectin receptor kinase III.1 |
4.593 |
0.314 |
| 2 |
AT3G43154 |
AT3G43154
|
[pseudogene] |
4.131 |
no data |
| 3 |
AT2G29250 |
AT2G29250
|
LecRK-III.2, L-type lectin receptor kinase III.2 |
4.086 |
0.116 |
| 4 |
AT5G48850 |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
3.869 |
0.224 |
| 5 |
AT3G30703 |
AT3G30703
|
[gypsy-like retrotransposon family, has a 6.1e-20 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
3.828 |
no data |
| 6 |
AT3G56710 |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
3.529 |
0.226 |
| 7 |
AT5G26220 |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
3.449 |
0.175 |
| 8 |
AT4G22485 |
AT4G22485
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
3.445 |
no data |
| 9 |
AT5G32161 |
AT5G32161
|
unknown |
3.386 |
no data |
| 10 |
AT1G47400 |
AT1G47400
|
unknown |
3.329 |
0.103 |
| 11 |
AT1G47395 |
AT1G47395
|
unknown |
3.269 |
no data |
| 12 |
AT5G32143 |
AT5G32143
|
[pseudogene] |
3.206 |
no data |
| 13 |
AT5G53380 |
AT5G53380
|
[O-acyltransferase (WSD1-like) family protein] |
3.206 |
no data |
| 14 |
AT4G03600 |
AT4G03600
|
unknown |
3.206 |
0.070 |
| 15 |
AT5G38910 |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
3.140 |
0.052 |
| 16 |
AT4G19750 |
AT4G19750
|
[Glycosyl hydrolase family protein with chitinase insertion domain] |
2.947 |
no data |
| 17 |
AT5G09430 |
AT5G09430
|
[alpha/beta-Hydrolases superfamily protein] |
2.920 |
-0.195 |
| 18 |
AT1G48460 |
AT1G48460
|
unknown |
2.920 |
-0.041 |
| 19 |
AT1G68120 |
AT1G68120
|
ATBPC3, BPC3, basic pentacysteine 3 |
2.855 |
0.089 |
| 20 |
AT2G25130 |
AT2G25130
|
[ARM repeat superfamily protein] |
2.835 |
no data |
| 21 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
2.808 |
no data |
| 22 |
AT2G27180 |
AT2G27180
|
unknown |
2.775 |
0.193 |
| 23 |
AT2G13810 |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
2.727 |
0.185 |
| 24 |
AT1G21525 |
AT1G21525
|
[expressed protein] |
2.715 |
no data |
| 25 |
AT1G01680 |
AT1G01680
|
ATPUB54, PLANT U-BOX 54, PUB54, plant U-box 54 |
2.669 |
no data |
| 26 |
AT1G35210 |
AT1G35210
|
unknown |
2.569 |
0.159 |
| 27 |
AT2G44800 |
AT2G44800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
2.538 |
-0.062 |
| 28 |
AT5G39180 |
AT5G39180
|
[RmlC-like cupins superfamily protein] |
2.518 |
no data |
| 29 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
2.505 |
0.077 |
| 30 |
AT2G02580 |
AT2G02580
|
CYP71B9, cytochrome P450, family 71, subfamily B, polypeptide 9 |
2.501 |
0.011 |
| 31 |
AT2G30890 |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
2.468 |
0.206 |
| 32 |
AT3G50845 |
AT3G50845
|
unknown |
2.465 |
no data |
| 33 |
AT5G65030 |
AT5G65030
|
unknown |
2.445 |
no data |
| 34 |
AT2G03821 |
AT2G03821
|
unknown |
2.410 |
no data |
| 35 |
AT2G39010 |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
2.410 |
0.200 |
| 36 |
AT5G07215 |
AT5G07215
|
[non-LTR retrotransposon family (LINE), has a 6.9e-34 P-value blast match to GB:NP_038605 L1 repeat, Tf subfamily, member 30 (LINE-element) (Mus musculus)] |
2.373 |
no data |
| 37 |
AT5G24180 |
AT5G24180
|
[Lipase class 3-related protein] |
2.325 |
no data |
| 38 |
AT1G45015 |
AT1G45015
|
[MD-2-related lipid recognition domain-containing protein] |
2.325 |
no data |
| 39 |
AT1G10680 |
AT1G10680
|
ABCB10, ATP-binding cassette B10, PGP10, P-glycoprotein 10 |
2.308 |
0.182 |
| 40 |
AT1G76952 |
AT1G76952
|
IDL5, inflorescence deficient in abscission (IDA)-like 5 |
2.286 |
no data |
| 41 |
AT1G19050 |
AT1G19050
|
ARR7, response regulator 7 |
2.250 |
0.128 |
| 42 |
AT3G05080 |
AT3G05080
|
unknown |
2.219 |
0.055 |
| 43 |
AT4G39364 |
AT4G39364
|
[snoRNA] |
2.219 |
no data |
| 44 |
AT2G02690 |
AT2G02690
|
[Cysteine/Histidine-rich C1 domain family protein] |
2.218 |
no data |
| 45 |
AT3G16220 |
AT3G16220
|
[Predicted eukaryotic LigT] |
2.217 |
0.171 |
| 46 |
AT3G55090 |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
2.182 |
0.160 |
| 47 |
AT3G59930 |
AT3G59930
|
unknown |
2.111 |
no data |
| 48 |
AT3G13677 |
AT3G13677
|
unknown |
2.050 |
no data |
| 49 |
AT3G50770 |
AT3G50770
|
CML41, calmodulin-like 41 |
2.049 |
0.396 |
| 50 |
AT2G19440 |
AT2G19440
|
[O-Glycosyl hydrolases family 17 protein] |
2.044 |
no data |
| 51 |
AT4G39361 |
AT4G39361
|
[snoRNA] |
2.042 |
no data |
| 52 |
AT2G04038 |
AT2G04038
|
AtbZIP48, basic leucine-zipper 48, bZIP48, basic leucine-zipper 48 |
2.021 |
-0.043 |
| 53 |
AT3G46230 |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
2.002 |
0.081 |
| 54 |
AT5G59360 |
AT5G59360
|
unknown |
1.967 |
-0.030 |
| 55 |
AT2G35550 |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
1.967 |
0.120 |
| 56 |
AT1G20070 |
AT1G20070
|
unknown |
1.937 |
0.162 |
| 57 |
AT5G26690 |
AT5G26690
|
[Heavy metal transport/detoxification superfamily protein ] |
1.933 |
no data |
| 58 |
AT2G34530 |
AT2G34530
|
unknown |
1.931 |
0.061 |
| 59 |
AT5G13320 |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
1.914 |
0.288 |
| 60 |
AT1G05420 |
AT1G05420
|
ATOFP12, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 12, OFP12, ovate family protein 12 |
1.911 |
no data |
| 61 |
AT3G53860 |
AT3G53860
|
unknown |
1.908 |
no data |
| 62 |
AT3G03910 |
AT3G03910
|
glutamate dehydrogenase 3 |
1.908 |
0.162 |
| 63 |
AT5G62330 |
AT5G62330
|
unknown |
1.905 |
0.069 |
| 64 |
AT3G28510 |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.885 |
0.037 |
| 65 |
AT3G48920 |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
1.869 |
0.031 |
| 66 |
AT1G10460 |
AT1G10460
|
GLP7, germin-like protein 7 |
1.869 |
0.114 |
| 67 |
AT1G02450 |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
1.866 |
0.152 |
| 68 |
AT1G48690 |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
1.863 |
0.134 |
| 69 |
AT4G13220 |
AT4G13220
|
unknown |
1.858 |
-0.011 |
| 70 |
AT5G52700 |
AT5G52700
|
[Copper transport protein family] |
1.826 |
no data |
| 71 |
AT3G53980 |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.821 |
-0.217 |
| 72 |
AT5G48780 |
AT5G48780
|
[disease resistance protein (TIR-NBS class)] |
1.817 |
0.144 |
| 73 |
AT4G32175 |
AT4G32175
|
[PNAS-3 related] |
1.806 |
no data |
| 74 |
AT2G28850 |
AT2G28850
|
CYP710A3, cytochrome P450, family 710, subfamily A, polypeptide 3 |
1.794 |
no data |
| 75 |
AT3G13240 |
AT3G13240
|
unknown |
1.780 |
-0.238 |
| 76 |
AT1G05220 |
AT1G05220
|
[Transmembrane protein 97, predicted] |
1.770 |
no data |
| 77 |
AT2G35750 |
AT2G35750
|
unknown |
1.770 |
-0.001 |
| 78 |
AT3G23530 |
AT3G23530
|
[Cyclopropane-fatty-acyl-phospholipid synthase] |
1.762 |
no data |
| 79 |
AT2G42940 |
AT2G42940
|
AHL16, AT-hook motif nuclear-localized protein 16, TEK, TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK |
1.757 |
-0.011 |
| 80 |
AT3G63052 |
AT3G63052
|
unknown |
1.757 |
no data |
| 81 |
AT2G14440 |
AT2G14440
|
[Leucine-rich repeat protein kinase family protein] |
1.722 |
no data |
| 82 |
AT1G68360 |
AT1G68360
|
[C2H2 and C2HC zinc fingers superfamily protein] |
1.721 |
0.049 |
| 83 |
AT5G27440 |
AT5G27440
|
unknown |
1.718 |
0.105 |
| 84 |
AT2G21210 |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
1.715 |
-0.020 |
| 85 |
AT4G25300 |
AT4G25300
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.713 |
no data |
| 86 |
AT3G23805 |
AT3G23805
|
RALFL24, ralf-like 24 |
1.665 |
0.112 |
| 87 |
AT1G21520 |
AT1G21520
|
unknown |
1.661 |
no data |
| 88 |
AT2G32860 |
AT2G32860
|
BGLU33, beta glucosidase 33 |
1.647 |
0.143 |
| 89 |
AT2G23960 |
AT2G23960
|
[Class I glutamine amidotransferase-like superfamily protein] |
1.623 |
0.344 |
| 90 |
AT5G41460 |
AT5G41460
|
unknown |
1.623 |
0.175 |
| 91 |
AT2G43920 |
AT2G43920
|
ATHOL2, HOL2, HARMLESS TO OZONE LAYER 2 |
1.618 |
no data |
| 92 |
AT4G10500 |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.602 |
0.594 |
| 93 |
AT5G66800 |
AT5G66800
|
unknown |
1.594 |
0.089 |
| 94 |
AT1G19390 |
AT1G19390
|
[Wall-associated kinase family protein] |
1.586 |
0.138 |
| 95 |
AT5G22930 |
AT5G22930
|
unknown |
1.581 |
no data |
| 96 |
AT2G36305 |
AT2G36305
|
ATFACE-2, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN-CONVERTING ENZYME-2, ATFACE2, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN-CONVERTING ENZYME 2, FACE2, farnesylated protein-converting enzyme 2, RCE1, RAS-CONVERTING ENZYME 1 |
1.580 |
no data |
| 97 |
AT5G15030 |
AT5G15030
|
[Paired amphipathic helix (PAH2) superfamily protein] |
1.562 |
no data |
| 98 |
AT5G41880 |
AT5G41880
|
POLA3, POLA4 |
1.562 |
-0.058 |
| 99 |
AT2G14860 |
AT2G14860
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
1.562 |
0.312 |
| 100 |
AT4G26560 |
AT4G26560
|
CBL7, calcineurin B-like protein 7 |
1.556 |
0.197 |
| 101 |
AT3G56730 |
AT3G56730
|
[Putative endonuclease or glycosyl hydrolase] |
1.543 |
-0.147 |
| 102 |
AT2G04110 |
AT2G04110
|
[expressed protein] |
1.541 |
no data |
| 103 |
AT3G01890 |
AT3G01890
|
[SWIB/MDM2 domain superfamily protein] |
1.528 |
0.136 |
| 104 |
AT1G44030 |
AT1G44030
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.519 |
0.054 |
| 105 |
AT1G01190 |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
1.515 |
0.290 |
| 106 |
AT3G10090 |
AT3G10090
|
[Nucleic acid-binding, OB-fold-like protein] |
1.506 |
-0.037 |
| 107 |
AT3G54070 |
AT3G54070
|
[Ankyrin repeat family protein] |
1.500 |
0.103 |
| 108 |
AT4G22790 |
AT4G22790
|
[MATE efflux family protein] |
1.494 |
0.180 |
| 109 |
AT1G67035 |
AT1G67035
|
unknown |
1.492 |
no data |
| 110 |
AT5G47455 |
AT5G47455
|
unknown |
1.491 |
-0.036 |
| 111 |
AT1G31320 |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
1.491 |
0.043 |
| 112 |
AT1G03920 |
AT1G03920
|
[Protein kinase family protein] |
1.490 |
-0.125 |
| 113 |
AT4G15990 |
AT4G15990
|
unknown |
1.475 |
no data |
| 114 |
AT3G48425 |
AT3G48425
|
[DNAse I-like superfamily protein] |
1.468 |
-0.041 |
| 115 |
AT3G24400 |
AT3G24400
|
AtPERK2, proline-rich extensin-like receptor kinase 2, PERK2, proline-rich extensin-like receptor kinase 2 |
1.463 |
-0.049 |
| 116 |
AT4G17970 |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
1.458 |
0.180 |
| 117 |
AT4G17340 |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
1.456 |
-0.021 |
| 118 |
AT2G15970 |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
1.451 |
-0.041 |
| 119 |
AT1G24280 |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
1.439 |
-0.249 |
| 120 |
AT3G50700 |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
1.435 |
-0.095 |
| 121 |
AT3G14560 |
AT3G14560
|
unknown |
1.419 |
-0.149 |
| 122 |
AT2G34610 |
AT2G34610
|
unknown |
1.412 |
0.117 |
| 123 |
AT5G58770 |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
1.408 |
-0.206 |
| 124 |
AT5G58784 |
AT5G58784
|
[Undecaprenyl pyrophosphate synthetase family protein] |
1.406 |
0.122 |
| 125 |
AT2G42360 |
AT2G42360
|
[RING/U-box superfamily protein] |
1.393 |
0.204 |
| 126 |
AT4G24430 |
AT4G24430
|
[Rhamnogalacturonate lyase family protein] |
1.393 |
0.131 |
| 127 |
AT1G76430 |
AT1G76430
|
PHT1;9, phosphate transporter 1;9 |
1.366 |
-0.015 |
| 128 |
AT3G43890 |
AT3G43890
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.360 |
no data |
| 129 |
AT5G18780 |
AT5G18780
|
[F-box/RNI-like superfamily protein] |
1.357 |
0.046 |
| 130 |
AT1G65850 |
AT1G65850
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
1.356 |
0.121 |
| 131 |
AT3G25882 |
AT3G25882
|
NIMIN-2, NIM1-interacting 2 |
1.350 |
no data |
| 132 |
AT3G24810 |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
1.342 |
0.299 |
| 133 |
AT5G53045 |
AT5G53045
|
unknown |
1.340 |
no data |
| 134 |
AT2G21900 |
AT2G21900
|
ATWRKY59, WRKY59, WRKY DNA-binding protein 59 |
1.340 |
no data |
| 135 |
AT1G64625 |
AT1G64625
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
1.340 |
no data |
| 136 |
AT5G50800 |
AT5G50800
|
AtSWEET13, SWEET13 |
1.332 |
0.175 |
| 137 |
AT5G17450 |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
1.329 |
-0.082 |
| 138 |
AT5G36870 |
AT5G36870
|
ATGSL09, glucan synthase-like 9, atgsl9, gsl09, GSL09, glucan synthase-like 9 |
1.327 |
0.118 |
| 139 |
AT3G50160 |
AT3G50160
|
unknown |
1.323 |
0.102 |
| 140 |
AT3G27070 |
AT3G27070
|
TOM20-1, translocase outer membrane 20-1 |
1.318 |
-0.084 |
| 141 |
AT4G27050 |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
1.317 |
0.037 |
| 142 |
AT5G60860 |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
1.316 |
-0.010 |
| 143 |
AT4G09420 |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
1.308 |
0.157 |
| 144 |
AT2G25410 |
AT2G25410
|
[RING/U-box superfamily protein] |
1.307 |
no data |
| 145 |
AT3G17180 |
AT3G17180
|
scpl33, serine carboxypeptidase-like 33 |
1.300 |
0.086 |
| 146 |
AT4G18630 |
AT4G18630
|
unknown |
1.292 |
0.119 |
| 147 |
AT3G45870 |
AT3G45870
|
UMAMIT3, Usually multiple acids move in and out Transporters 3 |
1.286 |
-0.284 |
| 148 |
AT1G58170 |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
1.282 |
0.198 |
| 149 |
AT3G53830 |
AT3G53830
|
[Regulator of chromosome condensation (RCC1) family protein] |
1.278 |
no data |
| 150 |
AT5G04160 |
AT5G04160
|
[Nucleotide-sugar transporter family protein] |
1.261 |
0.124 |
| 151 |
AT2G32930 |
AT2G32930
|
ZFN2, zinc finger nuclease 2 |
1.258 |
-0.011 |
| 152 |
AT1G52620 |
AT1G52620
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
1.257 |
-0.091 |
| 153 |
AT3G56430 |
AT3G56430
|
unknown |
1.255 |
0.008 |
| 154 |
AT4G10310 |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
1.252 |
0.174 |
| 155 |
AT2G20500 |
AT2G20500
|
unknown |
1.246 |
-0.017 |
| 156 |
AT5G09978 |
AT5G09978
|
PEP7, elicitor peptide 7 precursor |
1.242 |
no data |
| 157 |
AT3G27060 |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
1.240 |
0.010 |
| 158 |
AT1G18970 |
AT1G18970
|
GLP4, germin-like protein 4 |
1.228 |
0.131 |
| 159 |
AT3G18777 |
AT3G18777
|
[pseudogene] |
1.222 |
no data |
| 160 |
AT1G20290 |
AT1G20290
|
[SWI-SNF-related chromatin binding protein] |
1.222 |
0.030 |
| 161 |
AT4G10280 |
AT4G10280
|
[RmlC-like cupins superfamily protein] |
1.220 |
-0.042 |
| 162 |
AT2G16385 |
AT2G16385
|
unknown |
1.220 |
no data |
| 163 |
AT5G25451 |
AT5G25451
|
|
1.219 |
no data |
| 164 |
AT3G29780 |
AT3G29780
|
RALFL27, ralf-like 27 |
1.219 |
0.219 |
| 165 |
AT3G12580 |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
1.218 |
-0.029 |
| 166 |
AT3G14200 |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
1.217 |
0.079 |
| 167 |
AT5G08640 |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
1.211 |
-0.129 |
| 168 |
AT4G37630 |
AT4G37630
|
CYCD5;1, cyclin d5;1 |
1.208 |
0.115 |
| 169 |
AT5G39150 |
AT5G39150
|
[RmlC-like cupins superfamily protein] |
1.207 |
no data |
| 170 |
AT4G03610 |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
1.198 |
0.045 |
| 171 |
AT5G67230 |
AT5G67230
|
I14H, IRREGULAR XYLEM 14 Homolog, IRX14-L, IRREGULAR XYLEM 14-LIKE |
1.193 |
0.098 |
| 172 |
AT3G14540 |
AT3G14540
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
1.193 |
no data |
| 173 |
AT2G41720 |
AT2G41720
|
EMB2654, EMBRYO DEFECTIVE 2654 |
1.188 |
0.141 |
| 174 |
AT1G13910 |
AT1G13910
|
[Leucine-rich repeat (LRR) family protein] |
1.184 |
-0.043 |
| 175 |
AT5G39120 |
AT5G39120
|
[RmlC-like cupins superfamily protein] |
1.183 |
no data |
| 176 |
AT1G75010 |
AT1G75010
|
ARC3, ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 |
1.183 |
-0.208 |
| 177 |
AT1G19960 |
AT1G19960
|
unknown |
1.181 |
0.140 |
| 178 |
AT4G14390 |
AT4G14390
|
[Ankyrin repeat family protein] |
1.180 |
0.110 |
| 179 |
AT1G73600 |
AT1G73600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.178 |
no data |
| 180 |
AT1G73602 |
AT1G73602
|
CPuORF32, conserved peptide upstream open reading frame 32 |
1.177 |
no data |
| 181 |
AT5G03890 |
AT5G03890
|
unknown |
1.174 |
no data |
| 182 |
AT3G26775 |
AT3G26775
|
[pseudogene] |
1.173 |
no data |
| 183 |
AT4G21990 |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
1.165 |
0.047 |
| 184 |
AT5G66670 |
AT5G66670
|
unknown |
1.163 |
-0.158 |
| 185 |
AT4G00880 |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
1.161 |
0.075 |
| 186 |
AT4G30662 |
AT4G30662
|
unknown |
1.158 |
no data |
| 187 |
AT5G18930 |
AT5G18930
|
BUD2, BUSHY AND DWARF 2, SAMDC4 |
1.154 |
0.083 |
| 188 |
AT2G17060 |
AT2G17060
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
1.154 |
0.002 |
| 189 |
AT1G04770 |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
1.149 |
0.037 |
| 190 |
AT3G50100 |
AT3G50100
|
SDN1, small RNA degrading nuclease 1 |
1.146 |
no data |
| 191 |
AT1G74458 |
AT1G74458
|
unknown |
1.145 |
no data |
| 192 |
AT2G32940 |
AT2G32940
|
AGO6, ARGONAUTE 6 |
1.143 |
no data |
| 193 |
AT1G45120 |
AT1G45120
|
[copia-like retrotransposon family, has a 1.5e-294 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
1.139 |
no data |
| 194 |
AT3G21420 |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
1.137 |
0.032 |
| 195 |
AT3G21220 |
AT3G21220
|
ATMAP2K_ALPHA, ATMEK5, ARABIDOPSIS THALIANA MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5, ATMKK5, ARABIDOPSIS THALIANA MITOGEN-ACTIVATED PROTEIN KINASE KINASE 5, MAP2K_A, MEK5, MAP KINASE KINASE 5, MKK5, MAP kinase kinase 5 |
1.136 |
0.035 |
| 196 |
AT5G10110 |
AT5G10110
|
unknown |
1.132 |
no data |
| 197 |
AT3G03500 |
AT3G03500
|
[TatD related DNase] |
1.131 |
no data |
| 198 |
AT5G38100 |
AT5G38100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.130 |
no data |
| 199 |
AT2G40390 |
AT2G40390
|
unknown |
1.129 |
no data |
| 200 |
AT1G23340 |
AT1G23340
|
unknown |
1.127 |
0.062 |
| 201 |
AT1G66450 |
AT1G66450
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.125 |
0.045 |
| 202 |
AT4G09890 |
AT4G09890
|
unknown |
1.120 |
-0.050 |
| 203 |
AT5G10130 |
AT5G10130
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.117 |
0.179 |
| 204 |
AT1G53345 |
AT1G53345
|
unknown |
1.117 |
-0.004 |
| 205 |
AT2G25640 |
AT2G25640
|
[SPOC domain / Transcription elongation factor S-II protein] |
1.112 |
0.130 |
| 206 |
AT3G07900 |
AT3G07900
|
[O-fucosyltransferase family protein] |
1.093 |
-0.218 |
| 207 |
AT1G19715 |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
1.093 |
-0.159 |
| 208 |
AT5G05270 |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
1.087 |
0.031 |
| 209 |
AT2G27970 |
AT2G27970
|
CKS2, CDK-subunit 2 |
1.085 |
-0.001 |
| 210 |
AT4G39770 |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
1.084 |
0.150 |
| 211 |
AT1G73470 |
AT1G73470
|
unknown |
1.083 |
-0.010 |
| 212 |
AT2G48080 |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
1.080 |
0.312 |
| 213 |
AT4G16920 |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
1.074 |
0.073 |
| 214 |
AT1G62250 |
AT1G62250
|
unknown |
1.071 |
0.087 |
| 215 |
AT5G44110 |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
1.067 |
-0.302 |
| 216 |
AT5G26230 |
AT5G26230
|
MAKR1, membrane-associated kinase regulator 1 |
1.063 |
0.147 |
| 217 |
AT3G51470 |
AT3G51470
|
[Protein phosphatase 2C family protein] |
1.063 |
0.090 |
| 218 |
AT5G54710 |
AT5G54710
|
[Ankyrin repeat family protein] |
1.061 |
no data |
| 219 |
AT4G08685 |
AT4G08685
|
SAH7 |
1.058 |
0.136 |
| 220 |
AT3G20590 |
AT3G20590
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.055 |
no data |
| 221 |
AT5G25760 |
AT5G25760
|
PEX4, peroxin4, UBC21, ubiquitin-conjugating enzyme 21 |
1.053 |
0.010 |
| 222 |
AT5G02560 |
AT5G02560
|
HTA12, histone H2A 12 |
1.048 |
0.053 |
| 223 |
AT5G19370 |
AT5G19370
|
[rhodanese-like domain-containing protein / PPIC-type PPIASE domain-containing protein] |
1.046 |
-0.167 |
| 224 |
AT2G31530 |
AT2G31530
|
EMB2289, EMBRYO DEFECTIVE 2289, SCY2, SECY HOMOLOG 2 |
1.045 |
-0.130 |
| 225 |
AT1G30960 |
AT1G30960
|
[GTP-binding family protein] |
1.045 |
0.138 |
| 226 |
AT3G04770 |
AT3G04770
|
RPSAb, 40s ribosomal protein SA B |
1.043 |
0.035 |
| 227 |
AT4G37160 |
AT4G37160
|
sks15, SKU5 similar 15 |
1.043 |
0.067 |
| 228 |
AT2G02630 |
AT2G02630
|
[Cysteine/Histidine-rich C1 domain family protein] |
1.039 |
0.653 |
| 229 |
AT3G60520 |
AT3G60520
|
unknown |
1.039 |
0.007 |
| 230 |
AT1G51470 |
AT1G51470
|
BGLU35, beta glucosidase 35, TGG5, THIOGLUCOSIDE GLUCOHYDROLASE 5 |
1.032 |
no data |
| 231 |
AT2G33790 |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
1.032 |
0.025 |
| 232 |
AT5G22555 |
AT5G22555
|
unknown |
1.029 |
0.026 |
| 233 |
AT1G36370 |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
1.027 |
0.111 |
| 234 |
AT4G21740 |
AT4G21740
|
unknown |
1.022 |
no data |
| 235 |
AT4G22850 |
AT4G22850
|
[SNARE associated Golgi protein family] |
1.022 |
0.042 |
| 236 |
AT1G10480 |
AT1G10480
|
ZFP5, zinc finger protein 5 |
1.017 |
-0.050 |
| 237 |
AT2G40550 |
AT2G40550
|
ETG1, E2F target gene 1 |
1.016 |
-0.115 |
| 238 |
AT3G01750 |
AT3G01750
|
[Ankyrin repeat family protein] |
1.013 |
0.103 |
| 239 |
AT4G34930 |
AT4G34930
|
[PLC-like phosphodiesterases superfamily protein] |
1.011 |
no data |
| 240 |
AT3G29630 |
AT3G29630
|
[UDP-Glycosyltransferase superfamily protein] |
1.007 |
0.045 |
| 241 |
AT5G46260 |
AT5G46260
|
[disease resistance protein (TIR-NBS-LRR class) family] |
1.004 |
no data |
| 242 |
AT3G51240 |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.998 |
-0.577 |
| 243 |
AT3G51238 |
AT3G51238
|
other RNA |
0.998 |
no data |
| 244 |
AT5G58990 |
AT5G58990
|
unknown |
0.996 |
no data |
| 245 |
AT4G21230 |
AT4G21230
|
CRK27, cysteine-rich RLK (RECEPTOR-like protein kinase) 27 |
0.995 |
-0.113 |
| 246 |
AT5G59860 |
AT5G59860
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.994 |
0.069 |
| 247 |
AT1G62180 |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
0.993 |
0.003 |
| 248 |
AT1G01080 |
AT1G01080
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.992 |
0.137 |
| 249 |
AT2G22122 |
AT2G22122
|
unknown |
0.989 |
no data |
| 250 |
AT4G16370 |
AT4G16370
|
oligopeptide transporter |
0.984 |
0.051 |
| 251 |
AT3G24820 |
AT3G24820
|
[BSD domain-containing protein] |
0.983 |
-0.027 |
| 252 |
AT1G56120 |
AT1G56120
|
[Leucine-rich repeat transmembrane protein kinase] |
0.983 |
no data |
| 253 |
AT4G23730 |
AT4G23730
|
[Galactose mutarotase-like superfamily protein] |
0.982 |
0.166 |
| 254 |
AT3G06460 |
AT3G06460
|
[GNS1/SUR4 membrane protein family] |
0.979 |
0.096 |
| 255 |
AT5G11640 |
AT5G11640
|
[Thioredoxin superfamily protein] |
0.979 |
0.177 |
| 256 |
AT1G61480 |
AT1G61480
|
[S-locus lectin protein kinase family protein] |
0.979 |
0.107 |
| 257 |
AT2G24720 |
AT2G24720
|
ATGLR2.2, glutamate receptor 2.2, GLR2.2, glutamate receptor 2.2 |
0.978 |
no data |
| 258 |
AT5G44860 |
AT5G44860
|
unknown |
0.973 |
0.059 |
| 259 |
AT1G13930 |
AT1G13930
|
unknown |
0.970 |
-0.185 |
| 260 |
AT2G01940 |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.970 |
0.158 |
| 261 |
AT5G50140 |
AT5G50140
|
[Ankyrin repeat family protein] |
0.968 |
0.069 |
| 262 |
AT2G03090 |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
0.968 |
0.080 |
| 263 |
AT1G78000 |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.967 |
0.041 |
| 264 |
AT5G54970 |
AT5G54970
|
unknown |
0.966 |
0.236 |
| 265 |
AT5G52350 |
AT5G52350
|
ATEXO70A3, exocyst subunit exo70 family protein A3, EXO70A3, exocyst subunit exo70 family protein A3 |
0.965 |
0.109 |
| 266 |
AT4G13890 |
AT4G13890
|
EDA36, EMBRYO SAC DEVELOPMENT ARREST 37, EDA37, EMBRYO SAC DEVELOPMENT ARREST 36, SHM5, SERINE HYDROXYMETHYLTRANSFERASE 5 |
0.965 |
no data |
| 267 |
AT2G28790 |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
0.964 |
0.139 |
| 268 |
AT4G36190 |
AT4G36190
|
[Serine carboxypeptidase S28 family protein] |
0.963 |
no data |
| 269 |
AT1G63295 |
AT1G63295
|
[Remorin family protein] |
0.959 |
no data |
| 270 |
AT2G32610 |
AT2G32610
|
ATCSLB01, cellulose synthase-like B1, ATCSLB1, CELLULOSE SYNTHASE LIKE B1, CSLB01, CELLULOSE SYNTHASE LIKE B1, CSLB01, cellulose synthase-like B1 |
0.959 |
0.454 |
| 271 |
AT1G51640 |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
0.957 |
0.054 |
| 272 |
AT3G50350 |
AT3G50350
|
unknown |
0.954 |
-0.026 |
| 273 |
AT3G63440 |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.954 |
-0.129 |
| 274 |
AT3G13980 |
AT3G13980
|
unknown |
0.953 |
0.135 |
| 275 |
AT1G80760 |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
0.953 |
0.034 |
| 276 |
AT3G44940 |
AT3G44940
|
unknown |
0.952 |
0.177 |
| 277 |
AT1G08090 |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.951 |
0.468 |
| 278 |
AT4G23550 |
AT4G23550
|
ATWRKY29, WRKY29 |
0.948 |
no data |
| 279 |
AT2G23630 |
AT2G23630
|
sks16, SKU5 similar 16 |
0.948 |
0.195 |
| 280 |
AT2G27330 |
AT2G27330
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.948 |
0.024 |
| 281 |
AT1G30550 |
AT1G30550
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.946 |
0.102 |
| 282 |
AT5G50410 |
AT5G50410
|
unknown |
0.945 |
-0.066 |
| 283 |
AT4G04745 |
AT4G04745
|
unknown |
0.944 |
no data |
| 284 |
AT2G32560 |
AT2G32560
|
[F-box family protein] |
0.943 |
-0.049 |
| 285 |
AT3G18773 |
AT3G18773
|
[RING/U-box superfamily protein] |
0.943 |
no data |
| 286 |
AT1G61990 |
AT1G61990
|
[Mitochondrial transcription termination factor family protein] |
0.942 |
-0.103 |
| 287 |
AT5G15948 |
AT5G15948
|
CPuORF10, conserved peptide upstream open reading frame 10 |
0.942 |
no data |
| 288 |
AT5G15950 |
AT5G15950
|
[Adenosylmethionine decarboxylase family protein] |
0.942 |
no data |
| 289 |
AT5G46640 |
AT5G46640
|
AHL8, AT-hook motif nuclear localized protein 8 |
0.941 |
-0.013 |
| 290 |
AT3G18000 |
AT3G18000
|
NMT1, N-METHYLTRANSFERASE 1, PEAMT, XPL1, XIPOTL 1 |
0.939 |
no data |
| 291 |
AT5G50200 |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.939 |
0.158 |
| 292 |
AT3G17998 |
AT3G17998
|
CPuORF30, conserved peptide upstream open reading frame 30 |
0.939 |
no data |
| 293 |
AT5G59050 |
AT5G59050
|
unknown |
0.936 |
0.143 |
| 294 |
AT4G12270 |
AT4G12270
|
[Copper amine oxidase family protein] |
0.936 |
0.110 |
| 295 |
AT3G61610 |
AT3G61610
|
[Galactose mutarotase-like superfamily protein] |
0.934 |
0.093 |
| 296 |
AT5G54040 |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.932 |
0.223 |
| 297 |
AT3G11320 |
AT3G11320
|
[Nucleotide-sugar transporter family protein] |
0.931 |
-0.289 |
| 298 |
AT4G03140 |
AT4G03140
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.930 |
-0.240 |
| 299 |
AT1G04220 |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.928 |
0.287 |
| 300 |
AT1G52905 |
AT1G52905
|
unknown |
0.927 |
no data |
| 301 |
AT5G59170 |
AT5G59170
|
[Proline-rich extensin-like family protein] |
-4.296 |
-0.085 |
| 302 |
AT1G73510 |
AT1G73510
|
unknown |
-3.800 |
0.012 |
| 303 |
AT1G09080 |
AT1G09080
|
BIP3, binding protein 3 |
-3.732 |
-0.160 |
| 304 |
AT2G47520 |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-3.704 |
-0.004 |
| 305 |
AT5G45230 |
AT5G45230
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-3.530 |
0.169 |
| 306 |
AT3G22830 |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
-3.499 |
-0.137 |
| 307 |
AT1G76640 |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
-3.416 |
0.205 |
| 308 |
AT4G31970 |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
-3.339 |
0.229 |
| 309 |
AT1G05680 |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-3.320 |
0.349 |
| 310 |
AT2G05441 |
AT2G05441
|
|
-3.319 |
no data |
| 311 |
AT1G12610 |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-3.275 |
0.051 |
| 312 |
AT3G20395 |
AT3G20395
|
[RING/U-box superfamily protein] |
-3.272 |
no data |
| 313 |
AT2G36590 |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-3.201 |
-0.048 |
| 314 |
AT3G19025 |
AT3G19025
|
[pseudogene] |
-3.197 |
no data |
| 315 |
AT1G06030 |
AT1G06030
|
[pfkB-like carbohydrate kinase family protein] |
-3.146 |
no data |
| 316 |
AT5G58630 |
AT5G58630
|
TRM31, TON1 Recruiting Motif 31 |
-3.120 |
-0.207 |
| 317 |
AT1G63030 |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
-3.059 |
0.411 |
| 318 |
AT5G24600 |
AT5G24600
|
unknown |
-3.052 |
0.210 |
| 319 |
AT2G20625 |
AT2G20625
|
unknown |
-3.042 |
0.161 |
| 320 |
AT2G23270 |
AT2G23270
|
unknown |
-3.037 |
0.256 |
| 321 |
AT1G21140 |
AT1G21140
|
[Vacuolar iron transporter (VIT) family protein] |
-2.983 |
0.241 |
| 322 |
AT2G31083 |
AT2G31083
|
AtCLE5, CLE5, CLAVATA3/ESR-RELATED 5 |
-2.953 |
no data |
| 323 |
AT1G73530 |
AT1G73530
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-2.928 |
0.156 |
| 324 |
AT4G19680 |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-2.863 |
-0.180 |
| 325 |
AT2G31100 |
AT2G31100
|
[alpha/beta-Hydrolases superfamily protein] |
-2.860 |
-0.117 |
| 326 |
AT3G29970 |
AT3G29970
|
[B12D protein] |
-2.850 |
0.172 |
| 327 |
AT5G10040 |
AT5G10040
|
unknown |
-2.832 |
-0.059 |
| 328 |
AT1G17420 |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
-2.813 |
0.226 |
| 329 |
AT4G01360 |
AT4G01360
|
BPS3, BYPASS 3 |
-2.813 |
no data |
| 330 |
AT5G57200 |
AT5G57200
|
[ENTH/ANTH/VHS superfamily protein] |
-2.810 |
0.171 |
| 331 |
AT5G63350 |
AT5G63350
|
unknown |
-2.810 |
no data |
| 332 |
AT1G52120 |
AT1G52120
|
[Mannose-binding lectin superfamily protein] |
-2.808 |
no data |
| 333 |
AT1G67100 |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
-2.784 |
0.089 |
| 334 |
AT4G24231 |
AT4G24231
|
unknown |
-2.742 |
no data |
| 335 |
AT4G24170 |
AT4G24170
|
[ATP binding microtubule motor family protein] |
-2.741 |
0.070 |
| 336 |
AT1G14120 |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.721 |
0.017 |
| 337 |
AT3G07490 |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
-2.681 |
0.191 |
| 338 |
AT3G01140 |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
-2.677 |
0.108 |
| 339 |
AT3G01142 |
AT3G01142
|
other RNA |
-2.677 |
no data |
| 340 |
AT5G45113 |
AT5G45113
|
[mitochondrial transcription termination factor-related / mTERF-related] |
-2.668 |
no data |
| 341 |
AT1G15002 |
AT1G15002
|
|
-2.668 |
no data |
| 342 |
AT4G04930 |
AT4G04930
|
DES-1-LIKE |
-2.591 |
0.176 |
| 343 |
AT2G04040 |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-2.562 |
-0.034 |
| 344 |
AT4G25490 |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-2.503 |
-0.026 |
| 345 |
AT2G39510 |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-2.483 |
0.174 |
| 346 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
-2.396 |
0.070 |
| 347 |
AT3G01600 |
AT3G01600
|
anac044, NAC domain containing protein 44, NAC044, NAC domain containing protein 44 |
-2.301 |
no data |
| 348 |
AT5G63450 |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
-2.300 |
-0.183 |
| 349 |
AT3G23420 |
AT3G23420
|
[F-box and associated interaction domains-containing protein] |
-2.282 |
no data |
| 350 |
AT1G29930 |
AT1G29930
|
AB140, CAB1, chlorophyll A/B binding protein 1, CAB140, CHLOROPHYLL A/B PROTEIN 140, LHCB1.3, LIGHT-HARVESTING CHLOROPHYLL A/B-PROTEIN 1.3 |
-2.243 |
no data |
| 351 |
AT3G61400 |
AT3G61400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.240 |
0.134 |
| 352 |
AT5G61440 |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-2.235 |
0.239 |
| 353 |
AT4G32480 |
AT4G32480
|
unknown |
-2.216 |
no data |
| 354 |
AT3G47720 |
AT3G47720
|
SRO4, similar to RCD one 4 |
-2.198 |
no data |
| 355 |
AT5G55150 |
AT5G55150
|
unknown |
-2.187 |
no data |
| 356 |
AT4G33560 |
AT4G33560
|
[Wound-responsive family protein] |
-2.184 |
0.191 |
| 357 |
AT4G02005 |
AT4G02005
|
other RNA |
-2.170 |
no data |
| 358 |
AT1G11210 |
AT1G11210
|
unknown |
-2.164 |
0.059 |
| 359 |
AT2G42220 |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-2.164 |
0.048 |
| 360 |
AT2G44850 |
AT2G44850
|
unknown |
-2.120 |
-0.086 |
| 361 |
AT1G69880 |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
-2.120 |
0.131 |
| 362 |
AT3G18870 |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
-2.115 |
0.110 |
| 363 |
AT1G45140 |
AT1G45140
|
[non-LTR retrotransposon family (LINE), has a 1.4e-37 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-2.114 |
no data |
| 364 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-2.112 |
0.228 |
| 365 |
AT2G15880 |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
-2.109 |
0.102 |
| 366 |
AT3G15450 |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-2.097 |
0.063 |
| 367 |
AT5G22920 |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-2.089 |
0.116 |
| 368 |
AT2G32210 |
AT2G32210
|
unknown |
-2.060 |
no data |
| 369 |
AT1G15040 |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-2.059 |
0.130 |
| 370 |
AT3G48510 |
AT3G48510
|
unknown |
-2.052 |
-0.261 |
| 371 |
AT3G49110 |
AT3G49110
|
ATPCA, PEROXIDASE CA, ATPRX33, PRX33, PEROXIDASE 33, PRXCA, peroxidase CA |
-2.042 |
no data |
| 372 |
AT2G38230 |
AT2G38230
|
ATPDX1.1, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.1, PDX1.1, pyridoxine biosynthesis 1.1 |
-2.034 |
no data |
| 373 |
AT1G25112 |
AT1G25112
|
unknown |
-2.014 |
no data |
| 374 |
AT4G19530 |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-2.014 |
0.218 |
| 375 |
AT4G36850 |
AT4G36850
|
[PQ-loop repeat family protein / transmembrane family protein] |
-2.004 |
no data |
| 376 |
AT1G33930 |
AT1G33930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-2.004 |
0.128 |
| 377 |
AT5G65690 |
AT5G65690
|
PCK2, phosphoenolpyruvate carboxykinase 2, PEPCK, PHOSPHOENOLPYRUVATE CARBOXYKINASE |
-2.002 |
no data |
| 378 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-1.996 |
0.229 |
| 379 |
AT2G15890 |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-1.995 |
-0.015 |
| 380 |
AT5G56544 |
AT5G56544
|
|
-1.993 |
no data |
| 381 |
AT1G80440 |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-1.984 |
-0.070 |
| 382 |
AT4G26260 |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-1.980 |
0.093 |
| 383 |
AT3G08040 |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
-1.980 |
0.123 |
| 384 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-1.975 |
0.009 |
| 385 |
AT4G31940 |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-1.973 |
0.026 |
| 386 |
AT2G18735 |
AT2G18735
|
other RNA |
-1.960 |
no data |
| 387 |
AT5G56080 |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-1.959 |
0.072 |
| 388 |
AT3G48520 |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-1.956 |
-0.241 |
| 389 |
AT1G34510 |
AT1G34510
|
[Peroxidase superfamily protein] |
-1.946 |
0.155 |
| 390 |
AT1G19210 |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-1.932 |
-0.262 |
| 391 |
AT1G33055 |
AT1G33055
|
unknown |
-1.926 |
0.007 |
| 392 |
AT5G20250 |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-1.924 |
0.087 |
| 393 |
AT4G25220 |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-1.918 |
-0.218 |
| 394 |
AT1G76800 |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-1.915 |
0.185 |
| 395 |
AT2G20670 |
AT2G20670
|
unknown |
-1.907 |
0.163 |
| 396 |
AT3G62740 |
AT3G62740
|
BGLU7, beta glucosidase 7 |
-1.904 |
0.278 |
| 397 |
AT5G19940 |
AT5G19940
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
-1.904 |
0.023 |
| 398 |
AT3G07720 |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
-1.896 |
-0.045 |
| 399 |
AT4G37370 |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-1.891 |
-0.302 |
| 400 |
AT1G42980 |
AT1G42980
|
[Actin-binding FH2 (formin homology 2) family protein] |
-1.890 |
no data |
| 401 |
AT5G36260 |
AT5G36260
|
[Eukaryotic aspartyl protease family protein] |
-1.880 |
0.214 |
| 402 |
AT1G18310 |
AT1G18310
|
[glycosyl hydrolase family 81 protein] |
-1.865 |
0.066 |
| 403 |
AT5G51860 |
AT5G51860
|
AGL72, AGAMOUS-like 72 |
-1.857 |
-0.140 |
| 404 |
AT2G38823 |
AT2G38823
|
unknown |
-1.840 |
no data |
| 405 |
AT2G34180 |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-1.840 |
0.315 |
| 406 |
AT3G22121 |
AT3G22121
|
other RNA |
-1.835 |
0.095 |
| 407 |
AT3G22120 |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-1.833 |
-0.149 |
| 408 |
AT5G06640 |
AT5G06640
|
EXT10, extensin 10 |
-1.829 |
0.321 |
| 409 |
AT3G43190 |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-1.828 |
-0.057 |
| 410 |
AT1G75620 |
AT1G75620
|
[glyoxal oxidase-related protein] |
-1.828 |
0.099 |
| 411 |
AT1G14185 |
AT1G14185
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-1.824 |
no data |
| 412 |
AT2G21010 |
AT2G21010
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-1.814 |
no data |
| 413 |
AT4G24110 |
AT4G24110
|
unknown |
-1.814 |
0.078 |
| 414 |
AT4G10340 |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-1.812 |
0.116 |
| 415 |
AT5G24313 |
AT5G24313
|
unknown |
-1.803 |
0.103 |
| 416 |
AT1G16850 |
AT1G16850
|
unknown |
-1.790 |
-0.137 |
| 417 |
AT1G64480 |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
-1.785 |
-0.203 |
| 418 |
AT5G04730 |
AT5G04730
|
[Ankyrin-repeat containing protein] |
-1.779 |
0.075 |
| 419 |
AT1G73120 |
AT1G73120
|
unknown |
-1.762 |
0.059 |
| 420 |
AT4G24040 |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-1.757 |
0.131 |
| 421 |
AT3G21720 |
AT3G21720
|
ICL, isocitrate lyase |
-1.751 |
-0.094 |
| 422 |
AT1G71970 |
AT1G71970
|
unknown |
-1.750 |
0.070 |
| 423 |
AT5G63160 |
AT5G63160
|
BT1, BTB and TAZ domain protein 1 |
-1.734 |
no data |
| 424 |
AT1G69890 |
AT1G69890
|
unknown |
-1.720 |
0.403 |
| 425 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-1.714 |
-0.100 |
| 426 |
AT3G47860 |
AT3G47860
|
CHL, chloroplastic lipocalin |
-1.711 |
-0.019 |
| 427 |
AT1G77120 |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-1.710 |
-0.005 |
| 428 |
AT3G52840 |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-1.703 |
0.245 |
| 429 |
AT1G77380 |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-1.701 |
0.020 |
| 430 |
AT3G25190 |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-1.694 |
0.162 |
| 431 |
AT2G33110 |
AT2G33110
|
ATVAMP723, vesicle-associated membrane protein 723, VAMP723, VAMP723, vesicle-associated membrane protein 723 |
-1.677 |
no data |
| 432 |
AT1G56300 |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
-1.670 |
0.079 |
| 433 |
AT3G53360 |
AT3G53360
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.665 |
0.110 |
| 434 |
AT1G25400 |
AT1G25400
|
unknown |
-1.663 |
0.160 |
| 435 |
AT1G76600 |
AT1G76600
|
unknown |
-1.652 |
0.154 |
| 436 |
AT4G37095 |
AT4G37095
|
unknown |
-1.650 |
no data |
| 437 |
AT3G54530 |
AT3G54530
|
unknown |
-1.649 |
no data |
| 438 |
AT2G23910 |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.638 |
-0.111 |
| 439 |
AT4G05049 |
AT4G05049
|
|
-1.629 |
no data |
| 440 |
AT4G27450 |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-1.612 |
0.015 |
| 441 |
AT2G24980 |
AT2G24980
|
EXT6, extensin 6 |
-1.612 |
0.008 |
| 442 |
AT5G59570 |
AT5G59570
|
BOA, BROTHER OF LUX ARRHYTHMO |
-1.603 |
no data |
| 443 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-1.594 |
0.133 |
| 444 |
AT4G19690 |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-1.583 |
0.066 |
| 445 |
AT5G05410 |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-1.581 |
0.047 |
| 446 |
AT5G06630 |
AT5G06630
|
[proline-rich extensin-like family protein] |
-1.580 |
0.156 |
| 447 |
AT1G61520 |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-1.562 |
-0.016 |
| 448 |
AT4G13540 |
AT4G13540
|
unknown |
-1.556 |
no data |
| 449 |
AT4G30320 |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.552 |
0.067 |
| 450 |
AT1G50750 |
AT1G50750
|
[Plant mobile domain protein family] |
-1.544 |
0.036 |
| 451 |
AT5G37260 |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-1.539 |
-0.214 |
| 452 |
AT1G19025 |
AT1G19025
|
[DNA repair metallo-beta-lactamase family protein] |
-1.534 |
0.146 |
| 453 |
AT3G56790 |
AT3G56790
|
[RNA splicing factor-related] |
-1.526 |
no data |
| 454 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
-1.524 |
0.113 |
| 455 |
AT4G20160 |
AT4G20160
|
[LOCATED IN: chloroplast] |
-1.519 |
0.104 |
| 456 |
AT2G31860 |
AT2G31860
|
[pseudogene] |
-1.518 |
0.110 |
| 457 |
AT1G01580 |
AT1G01580
|
ATFRO2, FERRIC REDUCTION OXIDASE 2, FRD1, FERRIC CHELATE REDUCTASE DEFECTIVE 1, FRO2, ferric reduction oxidase 2 |
-1.514 |
no data |
| 458 |
AT5G02780 |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-1.509 |
0.169 |
| 459 |
AT3G48360 |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-1.505 |
0.254 |
| 460 |
AT5G58680 |
AT5G58680
|
[ARM repeat superfamily protein] |
-1.492 |
0.113 |
| 461 |
AT4G18370 |
AT4G18370
|
DEG5, degradation of periplasmic proteins 5, DEGP5, DEGP PROTEASE 5, HHOA, PROTEASE HHOA PRECUSOR |
-1.483 |
0.022 |
| 462 |
AT1G55200 |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-1.479 |
-0.302 |
| 463 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-1.476 |
-0.103 |
| 464 |
AT3G54590 |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-1.476 |
-0.080 |
| 465 |
AT1G75030 |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
-1.469 |
0.088 |
| 466 |
AT2G04050 |
AT2G04050
|
[MATE efflux family protein] |
-1.469 |
-0.186 |
| 467 |
AT1G67365 |
AT1G67365
|
other RNA |
-1.467 |
no data |
| 468 |
AT1G69490 |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-1.467 |
0.157 |
| 469 |
AT4G01870 |
AT4G01870
|
[tolB protein-related] |
-1.466 |
0.016 |
| 470 |
AT1G43800 |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN-DESATURASE6 |
-1.463 |
0.120 |
| 471 |
AT5G01595 |
AT5G01595
|
other RNA |
-1.459 |
no data |
| 472 |
AT2G31085 |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
-1.457 |
0.112 |
| 473 |
AT1G14960 |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.452 |
-0.023 |
| 474 |
AT5G64110 |
AT5G64110
|
[Peroxidase superfamily protein] |
-1.450 |
-0.045 |
| 475 |
AT5G01600 |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-1.443 |
0.042 |
| 476 |
AT1G53163 |
AT1G53163
|
unknown |
-1.432 |
no data |
| 477 |
AT5G43450 |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.431 |
-0.508 |
| 478 |
AT1G50930 |
AT1G50930
|
unknown |
-1.430 |
0.158 |
| 479 |
AT2G41640 |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-1.423 |
0.138 |
| 480 |
AT2G41730 |
AT2G41730
|
unknown |
-1.419 |
-0.076 |
| 481 |
AT1G70290 |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-1.416 |
0.061 |
| 482 |
AT3G61060 |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-1.414 |
0.111 |
| 483 |
AT2G02710 |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-1.414 |
0.088 |
| 484 |
AT1G56240 |
AT1G56240
|
AtPP2-B13, phloem protein 2-B13, PP2-B13, phloem protein 2-B13 |
-1.410 |
no data |
| 485 |
AT1G56242 |
AT1G56242
|
other RNA |
-1.410 |
no data |
| 486 |
AT1G62500 |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.407 |
0.086 |
| 487 |
AT4G18280 |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-1.404 |
0.001 |
| 488 |
AT1G63180 |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-1.403 |
0.221 |
| 489 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-1.401 |
-0.042 |
| 490 |
AT1G59850 |
AT1G59850
|
[ARM repeat superfamily protein] |
-1.395 |
0.141 |
| 491 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
-1.394 |
0.069 |
| 492 |
AT1G34050 |
AT1G34050
|
[Ankyrin repeat family protein] |
-1.393 |
no data |
| 493 |
AT1G29640 |
AT1G29640
|
unknown |
-1.393 |
no data |
| 494 |
AT5G01380 |
AT5G01380
|
[Homeodomain-like superfamily protein] |
-1.391 |
0.145 |
| 495 |
AT3G03270 |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-1.390 |
0.048 |
| 496 |
AT5G54470 |
AT5G54470
|
BBX29, B-box domain protein 29 |
-1.388 |
-0.024 |
| 497 |
AT1G19530 |
AT1G19530
|
unknown |
-1.382 |
-0.035 |
| 498 |
AT2G19800 |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-1.380 |
0.179 |
| 499 |
AT2G44130 |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-1.375 |
0.193 |
| 500 |
AT3G58810 |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
-1.366 |
0.233 |
| 501 |
AT1G52690 |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-1.362 |
-0.141 |
| 502 |
AT4G37610 |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-1.362 |
0.173 |
| 503 |
AT3G07650 |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-1.362 |
0.127 |
| 504 |
AT4G36230 |
AT4G36230
|
unknown |
-1.360 |
0.124 |
| 505 |
AT1G56430 |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-1.359 |
0.079 |
| 506 |
AT5G04950 |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-1.358 |
0.007 |
| 507 |
AT2G39830 |
AT2G39830
|
DAR2, DA1-related protein 2, LRD3, LATERAL ROOT DEVELOPMENT 3 |
-1.345 |
0.179 |
| 508 |
AT2G32090 |
AT2G32090
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-1.337 |
-0.110 |
| 509 |
AT3G46240 |
AT3G46240
|
unknown |
-1.337 |
0.141 |
| 510 |
AT2G16890 |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
-1.335 |
0.690 |
| 511 |
AT1G74470 |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
-1.331 |
-0.122 |
| 512 |
AT2G18890 |
AT2G18890
|
[Protein kinase superfamily protein] |
-1.328 |
0.108 |
| 513 |
AT4G24710 |
AT4G24710
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.328 |
-0.030 |
| 514 |
AT4G38420 |
AT4G38420
|
sks9, SKU5 similar 9 |
-1.322 |
0.277 |
| 515 |
AT1G67980 |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-1.316 |
0.399 |
| 516 |
AT1G76220 |
AT1G76220
|
unknown |
-1.313 |
0.277 |
| 517 |
AT1G60190 |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-1.310 |
-0.100 |
| 518 |
AT5G18670 |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-1.309 |
0.184 |
| 519 |
AT5G06990 |
AT5G06990
|
unknown |
-1.306 |
no data |
| 520 |
AT5G22410 |
AT5G22410
|
RHS18, root hair specific 18 |
-1.297 |
-0.053 |
| 521 |
AT4G33070 |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-1.291 |
0.074 |
| 522 |
AT5G13210 |
AT5G13210
|
[Uncharacterised conserved protein UCP015417, vWA] |
-1.289 |
no data |
| 523 |
AT5G15970 |
AT5G15970
|
AtCor6.6, COR6.6, COLD-RESPONSIVE 6.6, KIN2 |
-1.282 |
no data |
| 524 |
AT1G67820 |
AT1G67820
|
[Protein phosphatase 2C family protein] |
-1.282 |
-0.049 |
| 525 |
AT5G66450 |
AT5G66450
|
LPPepsilon2, lipid phosphate phosphatase epsilon 2 |
-1.274 |
no data |
| 526 |
AT2G18700 |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-1.274 |
0.064 |
| 527 |
AT3G54580 |
AT3G54580
|
[Proline-rich extensin-like family protein] |
-1.267 |
-0.096 |
| 528 |
AT2G39400 |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-1.265 |
0.077 |
| 529 |
AT1G67105 |
AT1G67105
|
other RNA |
-1.262 |
no data |
| 530 |
AT5G15850 |
AT5G15850
|
ATCOL1, BBX2, B-box domain protein 2, COL1, CONSTANS-like 1 |
-1.261 |
0.006 |
| 531 |
AT5G15845 |
AT5G15845
|
other RNA |
-1.258 |
no data |
| 532 |
AT5G24770 |
AT5G24770
|
ATVSP2, VSP2, vegetative storage protein 2 |
-1.255 |
no data |
| 533 |
AT3G01970 |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
-1.253 |
0.072 |
| 534 |
AT4G25790 |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.252 |
0.072 |
| 535 |
AT5G11180 |
AT5G11180
|
ATGLR2.6, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.6, GLR2.6, glutamate receptor 2.6 |
-1.252 |
0.150 |
| 536 |
AT4G25410 |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.252 |
-0.109 |
| 537 |
AT2G18193 |
AT2G18193
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.251 |
no data |
| 538 |
AT4G17490 |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-1.251 |
0.079 |
| 539 |
AT5G67400 |
AT5G67400
|
RHS19, root hair specific 19 |
-1.247 |
0.116 |
| 540 |
AT4G28405 |
AT4G28405
|
[Expressed protein] |
-1.245 |
no data |
| 541 |
AT1G78830 |
AT1G78830
|
[Curculin-like (mannose-binding) lectin family protein] |
-1.245 |
no data |
| 542 |
AT3G30775 |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-1.240 |
0.050 |
| 543 |
AT3G52740 |
AT3G52740
|
unknown |
-1.234 |
0.117 |
| 544 |
AT1G61080 |
AT1G61080
|
[Hydroxyproline-rich glycoprotein family protein] |
-1.232 |
-0.015 |
| 545 |
AT4G37240 |
AT4G37240
|
unknown |
-1.231 |
0.247 |
| 546 |
AT1G19190 |
AT1G19190
|
[alpha/beta-Hydrolases superfamily protein] |
-1.231 |
-0.140 |
| 547 |
AT4G37780 |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
-1.230 |
0.077 |
| 548 |
AT5G02020 |
AT5G02020
|
SIS, Salt Induced Serine rich |
-1.219 |
0.048 |
| 549 |
AT3G07200 |
AT3G07200
|
[RING/U-box superfamily protein] |
-1.218 |
0.163 |
| 550 |
AT5G20830 |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
-1.212 |
0.092 |
| 551 |
AT1G74930 |
AT1G74930
|
ORA47 |
-1.212 |
0.138 |
| 552 |
AT3G14260 |
AT3G14260
|
unknown |
-1.211 |
no data |
| 553 |
AT3G16690 |
AT3G16690
|
AtSWEET16, SWEET16 |
-1.210 |
-0.010 |
| 554 |
AT4G25960 |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-1.205 |
0.257 |
| 555 |
AT1G26790 |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
-1.203 |
0.103 |
| 556 |
AT3G24520 |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-1.200 |
0.020 |
| 557 |
AT3G24518 |
AT3G24518
|
other RNA |
-1.199 |
no data |
| 558 |
AT1G18740 |
AT1G18740
|
unknown |
-1.196 |
-0.044 |
| 559 |
AT1G74450 |
AT1G74450
|
unknown |
-1.196 |
-0.070 |
| 560 |
AT2G20520 |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-1.195 |
0.144 |
| 561 |
AT4G24230 |
AT4G24230
|
ACBP3, acyl-CoA-binding domain 3 |
-1.191 |
no data |
| 562 |
AT2G30615 |
AT2G30615
|
unknown |
-1.190 |
no data |
| 563 |
AT2G42430 |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-1.190 |
-0.352 |
| 564 |
AT3G57760 |
AT3G57760
|
[Protein kinase superfamily protein] |
-1.188 |
-0.194 |
| 565 |
AT1G80380 |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.182 |
0.024 |
| 566 |
AT2G40000 |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-1.181 |
0.051 |
| 567 |
AT1G05250 |
AT1G05250
|
AtPRX1, PRX2, peroxidase 2 |
-1.178 |
no data |
| 568 |
AT4G33950 |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-1.178 |
-0.157 |
| 569 |
AT1G12244 |
AT1G12244
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-1.177 |
no data |
| 570 |
AT5G47220 |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-1.177 |
0.191 |
| 571 |
AT1G28100 |
AT1G28100
|
unknown |
-1.174 |
-0.008 |
| 572 |
AT1G56250 |
AT1G56250
|
AtPP2-B14, phloem protein 2-B14, PP2-B14, phloem protein 2-B14, VBF, VIP1-binding F-box protein |
-1.170 |
no data |
| 573 |
AT4G23700 |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-1.162 |
0.393 |
| 574 |
AT3G61160 |
AT3G61160
|
[Protein kinase superfamily protein] |
-1.159 |
0.041 |
| 575 |
AT5G46110 |
AT5G46110
|
APE2, ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT 2, TPT, triose-phosphate ⁄ phosphate translocator |
-1.152 |
-0.039 |
| 576 |
AT5G65207 |
AT5G65207
|
unknown |
-1.149 |
no data |
| 577 |
AT1G12805 |
AT1G12805
|
[nucleotide binding] |
-1.144 |
no data |
| 578 |
AT3G48180 |
AT3G48180
|
unknown |
-1.137 |
no data |
| 579 |
AT4G38770 |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-1.135 |
0.150 |
| 580 |
AT5G54270 |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-1.135 |
-0.026 |
| 581 |
AT1G01240 |
AT1G01240
|
unknown |
-1.132 |
0.155 |
| 582 |
AT3G12900 |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.129 |
0.140 |
| 583 |
AT2G25240 |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-1.125 |
-0.169 |
| 584 |
AT3G16350 |
AT3G16350
|
[Homeodomain-like superfamily protein] |
-1.124 |
-0.016 |
| 585 |
AT2G39310 |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-1.123 |
-0.339 |
| 586 |
AT2G34500 |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
-1.122 |
0.190 |
| 587 |
AT3G45700 |
AT3G45700
|
[Major facilitator superfamily protein] |
-1.121 |
0.145 |
| 588 |
AT5G61350 |
AT5G61350
|
[Protein kinase superfamily protein] |
-1.121 |
-0.052 |
| 589 |
AT2G15680 |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-1.119 |
0.047 |
| 590 |
AT1G04700 |
AT1G04700
|
[PB1 domain-containing protein tyrosine kinase] |
-1.119 |
0.258 |
| 591 |
AT4G13390 |
AT4G13390
|
EXT12, extensin 12 |
-1.119 |
0.173 |
| 592 |
AT4G38470 |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-1.117 |
0.139 |
| 593 |
AT5G39890 |
AT5G39890
|
unknown |
-1.116 |
0.078 |
| 594 |
AT3G48940 |
AT3G48940
|
[Remorin family protein] |
-1.116 |
0.005 |
| 595 |
AT5G17300 |
AT5G17300
|
RVE1, REVEILLE 1 |
-1.114 |
0.291 |
| 596 |
AT2G25090 |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-1.112 |
0.236 |
| 597 |
AT2G35150 |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
-1.112 |
0.266 |
| 598 |
AT4G29780 |
AT4G29780
|
unknown |
-1.108 |
0.155 |
| 599 |
AT1G70460 |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-1.104 |
0.008 |
| 600 |
AT3G48450 |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-1.103 |
0.501 |