|
probeID |
AGICode |
Annotation |
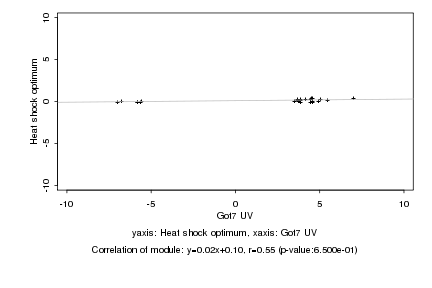
Log2 signal ratio
Got7 UV |
Log2 signal ratio
Heat shock optimum |
| 1 |
AT2G24850 |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
6.990 |
0.370 |
| 2 |
AT2G30770 |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
5.444 |
0.153 |
| 3 |
AT2G29350 |
AT2G29350
|
SAG13, senescence-associated gene 13 |
5.030 |
0.274 |
| 4 |
AT2G45220 |
AT2G45220
|
PME17, pectin methylesterase 17 |
4.887 |
0.068 |
| 5 |
AT1G33960 |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
4.567 |
0.018 |
| 6 |
AT2G13810 |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
4.548 |
-0.021 |
| 7 |
AT4G12480 |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
4.548 |
0.478 |
| 8 |
AT2G39030 |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
4.465 |
0.374 |
| 9 |
AT3G09940 |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
4.449 |
-0.013 |
| 10 |
AT1G73260 |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
4.446 |
0.335 |
| 11 |
AT3G25180 |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
4.106 |
0.301 |
| 12 |
AT2G38240 |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
3.821 |
0.185 |
| 13 |
AT4G16730 |
AT4G16730
|
TPS02, terpene synthase 02 |
3.821 |
-0.042 |
| 14 |
AT3G26830 |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
3.814 |
0.322 |
| 15 |
AT5G05340 |
AT5G05340
|
PRX52, peroxidase 52 |
3.690 |
0.041 |
| 16 |
AT3G50770 |
AT3G50770
|
CML41, calmodulin-like 41 |
3.654 |
0.135 |
| 17 |
AT3G55970 |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
3.639 |
0.249 |
| 18 |
AT3G21500 |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
3.464 |
0.041 |
| 19 |
AT3G01345 |
AT3G01345
|
[Expressed protein] |
-9.770 |
no data |
| 20 |
AT3G29739 |
AT3G29739
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 1.8e-33 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
-7.296 |
no data |
| 21 |
AT5G41660 |
AT5G41660
|
unknown |
-7.029 |
-0.001 |
| 22 |
AT1G72930 |
AT1G72930
|
AtTN10, TIR, toll/interleukin-1 receptor-like, TN10, TIR-nucleotide binding site family 10 |
-6.892 |
no data |
| 23 |
AT1G69140 |
AT1G69140
|
[pseudogene] |
-6.840 |
no data |
| 24 |
AT5G17890 |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-6.780 |
0.017 |
| 25 |
AT3G29734 |
AT3G29734
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 6.5e-179 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-6.621 |
no data |
| 26 |
AT2G31150 |
AT2G31150
|
[ATP binding] |
-6.582 |
no data |
| 27 |
AT3G29736 |
AT3G29736
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 7.4e-29 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-6.532 |
no data |
| 28 |
AT4G21640 |
AT4G21640
|
[Subtilase family protein] |
-6.370 |
no data |
| 29 |
AT3G47290 |
AT3G47290
|
ATPLC8, PLC8, phosphatidylinositol-speciwc phospholipase C8 |
-6.312 |
no data |
| 30 |
AT5G42825 |
AT5G42825
|
unknown |
-6.051 |
no data |
| 31 |
AT3G47250 |
AT3G47250
|
unknown |
-5.844 |
-0.025 |
| 32 |
AT1G35612 |
AT1G35612
|
[expressed protein] |
-5.821 |
-0.034 |
| 33 |
AT3G42052 |
AT3G42052
|
[gypsy-like retrotransposon family, has a 4.0e-162 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-5.777 |
no data |
| 34 |
AT5G17880 |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-5.660 |
-0.002 |
| 35 |
AT3G43740 |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-5.593 |
0.017 |