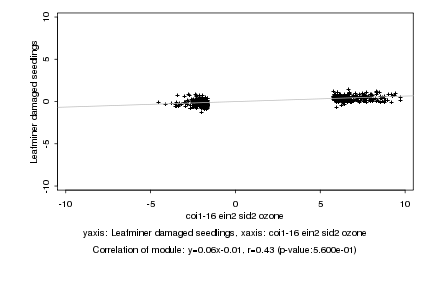
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
coi1-16 ein2 sid2 ozone |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
AT3G21520 |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
9.705 |
0.489 |
| 2 |
AT4G11170 |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
9.670 |
0.157 |
| 3 |
AT5G56960 |
AT5G56960
|
[basic helix-loop-helix (bHLH) DNA-binding family protein] |
9.590 |
no data |
| 4 |
AT1G79680 |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
9.405 |
1.058 |
| 5 |
AT1G26380 |
AT1G26380
|
[FAD-binding Berberine family protein] |
9.347 |
0.786 |
| 6 |
AT5G64905 |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
9.226 |
0.682 |
| 7 |
AT3G12910 |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
9.169 |
-0.001 |
| 8 |
AT3G02840 |
AT3G02840
|
[ARM repeat superfamily protein] |
9.140 |
0.864 |
| 9 |
AT2G30750 |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
9.013 |
0.852 |
| 10 |
AT4G31950 |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
8.932 |
0.130 |
| 11 |
AT1G21120 |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
8.913 |
0.186 |
| 12 |
AT4G23280 |
AT4G23280
|
CRK20, cysteine-rich RLK (RECEPTOR-like protein kinase) 20 |
8.829 |
0.222 |
| 13 |
AT3G54150 |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
8.757 |
0.693 |
| 14 |
AT5G46295 |
AT5G46295
|
unknown |
8.752 |
-0.011 |
| 15 |
AT1G14540 |
AT1G14540
|
PER4, peroxidase 4 |
8.730 |
0.476 |
| 16 |
AT5G05300 |
AT5G05300
|
unknown |
8.643 |
0.524 |
| 17 |
AT4G11470 |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
8.625 |
0.083 |
| 18 |
AT1G66600 |
AT1G66600
|
ABO3, ABA overly sensitive mutant 3, ATWRKY63, WRKY63, WRKY DNA-binding protein 63 |
8.600 |
no data |
| 19 |
AT2G02930 |
AT2G02930
|
ATGSTF3, glutathione S-transferase F3, GST16, GLUTATHIONE S-TRANSFERASE 16, GSTF3, glutathione S-transferase F3 |
8.577 |
no data |
| 20 |
AT2G37430 |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
8.560 |
0.468 |
| 21 |
AT5G24110 |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
8.520 |
0.445 |
| 22 |
AT1G51890 |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
8.509 |
0.650 |
| 23 |
AT2G29100 |
AT2G29100
|
ATGLR2.9, glutamate receptor 2.9, GLR2.9, GLUTAMATE RECEPTOR 2.9, GLR2.9, glutamate receptor 2.9 |
8.496 |
-0.006 |
| 24 |
AT5G61160 |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
8.469 |
1.075 |
| 25 |
AT1G08860 |
AT1G08860
|
BON3, BONZAI 3 |
8.379 |
0.022 |
| 26 |
AT1G30370 |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
8.352 |
0.140 |
| 27 |
AT2G26560 |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
8.340 |
1.122 |
| 28 |
AT1G05675 |
AT1G05675
|
[UDP-Glycosyltransferase superfamily protein] |
8.336 |
no data |
| 29 |
AT3G01830 |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
8.312 |
1.304 |
| 30 |
AT2G40740 |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
8.303 |
0.070 |
| 31 |
AT3G23250 |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
8.264 |
1.110 |
| 32 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
8.258 |
0.866 |
| 33 |
AT1G56060 |
AT1G56060
|
unknown |
8.250 |
0.414 |
| 34 |
AT4G15236 |
AT4G15236
|
ABCG43, ATP-binding cassette G43 |
8.208 |
no data |
| 35 |
AT1G21110 |
AT1G21110
|
IGMT3, indole glucosinolate O-methyltransferase 3 |
8.179 |
no data |
| 36 |
AT2G33710 |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
8.096 |
0.279 |
| 37 |
AT4G18540 |
AT4G18540
|
unknown |
8.038 |
0.024 |
| 38 |
AT5G36925 |
AT5G36925
|
unknown |
8.033 |
no data |
| 39 |
AT1G01560 |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
8.032 |
0.716 |
| 40 |
AT1G47890 |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
8.025 |
0.158 |
| 41 |
AT3G26830 |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
8.008 |
0.856 |
| 42 |
AT1G51920 |
AT1G51920
|
unknown |
7.998 |
0.364 |
| 43 |
AT1G52200 |
AT1G52200
|
[PLAC8 family protein] |
7.973 |
0.614 |
| 44 |
AT5G66890 |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
7.955 |
0.087 |
| 45 |
AT4G28460 |
AT4G28460
|
unknown |
7.943 |
0.082 |
| 46 |
AT5G47850 |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
7.939 |
0.284 |
| 47 |
AT1G26420 |
AT1G26420
|
[FAD-binding Berberine family protein] |
7.933 |
0.840 |
| 48 |
AT5G41730 |
AT5G41730
|
[Protein kinase family protein] |
7.924 |
0.183 |
| 49 |
AT1G33030 |
AT1G33030
|
[O-methyltransferase family protein] |
7.919 |
0.711 |
| 50 |
AT5G44990 |
AT5G44990
|
[Glutathione S-transferase family protein] |
7.903 |
0.211 |
| 51 |
AT5G08240 |
AT5G08240
|
unknown |
7.888 |
0.593 |
| 52 |
AT1G78410 |
AT1G78410
|
[VQ motif-containing protein] |
7.804 |
0.596 |
| 53 |
AT2G18690 |
AT2G18690
|
unknown |
7.767 |
0.560 |
| 54 |
AT1G02920 |
AT1G02920
|
ATGST11, ARABIDOPSIS GLUTATHIONE S-TRANSFERASE 11, ATGSTF7, ATGSTF8, GST11, GLUTATHIONE S-TRANSFERASE 11, GSTF7, glutathione S-transferase 7 |
7.734 |
no data |
| 55 |
AT4G04540 |
AT4G04540
|
CRK39, cysteine-rich RLK (RECEPTOR-like protein kinase) 39 |
7.731 |
no data |
| 56 |
AT1G70170 |
AT1G70170
|
MMP, matrix metalloproteinase |
7.722 |
0.224 |
| 57 |
AT1G71520 |
AT1G71520
|
[Integrase-type DNA-binding superfamily protein] |
7.720 |
no data |
| 58 |
AT1G71390 |
AT1G71390
|
AtRLP11, receptor like protein 11, RLP11, receptor like protein 11 |
7.719 |
0.002 |
| 59 |
AT4G22710 |
AT4G22710
|
CYP706A2, cytochrome P450, family 706, subfamily A, polypeptide 2 |
7.719 |
no data |
| 60 |
AT1G15520 |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
7.715 |
0.442 |
| 61 |
AT4G20000 |
AT4G20000
|
[VQ motif-containing protein] |
7.701 |
0.228 |
| 62 |
AT3G09940 |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
7.685 |
1.117 |
| 63 |
AT4G18430 |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
7.684 |
0.380 |
| 64 |
AT2G32200 |
AT2G32200
|
unknown |
7.662 |
no data |
| 65 |
AT1G49000 |
AT1G49000
|
unknown |
7.637 |
0.473 |
| 66 |
AT2G35980 |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
7.627 |
0.780 |
| 67 |
AT3G23550 |
AT3G23550
|
[MATE efflux family protein] |
7.625 |
0.685 |
| 68 |
AT1G19020 |
AT1G19020
|
unknown |
7.621 |
0.643 |
| 69 |
AT5G25910 |
AT5G25910
|
AtRLP52, receptor like protein 52, RLP52, receptor like protein 52 |
7.604 |
0.156 |
| 70 |
AT4G23215 |
AT4G23215
|
|
7.603 |
no data |
| 71 |
AT2G15390 |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
7.602 |
0.638 |
| 72 |
AT1G16420 |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
7.585 |
0.266 |
| 73 |
AT5G59530 |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
7.582 |
0.291 |
| 74 |
AT4G39670 |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
7.577 |
0.939 |
| 75 |
AT3G55150 |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
7.549 |
0.012 |
| 76 |
AT5G19880 |
AT5G19880
|
[Peroxidase superfamily protein] |
7.536 |
0.031 |
| 77 |
AT3G52400 |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
7.534 |
0.653 |
| 78 |
AT1G53625 |
AT1G53625
|
unknown |
7.533 |
no data |
| 79 |
AT5G20230 |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
7.525 |
0.806 |
| 80 |
AT1G30730 |
AT1G30730
|
[FAD-binding Berberine family protein] |
7.522 |
no data |
| 81 |
AT3G48630 |
AT3G48630
|
unknown |
7.507 |
no data |
| 82 |
AT1G66090 |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
7.506 |
0.592 |
| 83 |
AT5G11210 |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
7.497 |
0.387 |
| 84 |
AT2G39200 |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
7.494 |
0.610 |
| 85 |
AT5G65600 |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
7.493 |
0.611 |
| 86 |
AT5G44585 |
AT5G44585
|
unknown |
7.490 |
no data |
| 87 |
AT3G26500 |
AT3G26500
|
PIRL2, plant intracellular ras group-related LRR 2 |
7.461 |
no data |
| 88 |
AT1G32350 |
AT1G32350
|
AOX1D, alternative oxidase 1D |
7.452 |
0.987 |
| 89 |
AT1G51800 |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
7.432 |
0.650 |
| 90 |
AT5G22530 |
AT5G22530
|
unknown |
7.402 |
0.375 |
| 91 |
AT1G66700 |
AT1G66700
|
PXMT1 |
7.362 |
no data |
| 92 |
AT5G48657 |
AT5G48657
|
[defense protein-related] |
7.346 |
no data |
| 93 |
AT3G28580 |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
7.334 |
0.181 |
| 94 |
AT2G44370 |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
7.323 |
0.514 |
| 95 |
AT1G30700 |
AT1G30700
|
[FAD-binding Berberine family protein] |
7.279 |
0.935 |
| 96 |
AT4G37290 |
AT4G37290
|
unknown |
7.275 |
0.512 |
| 97 |
AT5G37490 |
AT5G37490
|
[ARM repeat superfamily protein] |
7.274 |
-0.010 |
| 98 |
AT4G14370 |
AT4G14370
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
7.268 |
no data |
| 99 |
AT1G14870 |
AT1G14870
|
AtPCR2, PCR2, PLANT CADMIUM RESISTANCE 2 |
7.230 |
no data |
| 100 |
AT4G04480 |
AT4G04480
|
unknown |
7.201 |
0.041 |
| 101 |
AT1G36600 |
AT1G36600
|
[copia-like retrotransposon family, has a 8.9e-21 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
7.195 |
0.050 |
| 102 |
AT2G25470 |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
7.186 |
0.467 |
| 103 |
AT5G02780 |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
7.185 |
0.594 |
| 104 |
AT5G35735 |
AT5G35735
|
[Auxin-responsive family protein] |
7.163 |
0.634 |
| 105 |
AT1G51820 |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
7.159 |
0.465 |
| 106 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
7.155 |
0.581 |
| 107 |
AT1G36622 |
AT1G36622
|
unknown |
7.124 |
no data |
| 108 |
AT1G59950 |
AT1G59950
|
[NAD(P)-linked oxidoreductase superfamily protein] |
7.112 |
0.354 |
| 109 |
AT5G57010 |
AT5G57010
|
[calmodulin-binding family protein] |
7.107 |
no data |
| 110 |
AT1G61560 |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
7.104 |
0.661 |
| 111 |
AT3G22600 |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
7.089 |
0.651 |
| 112 |
AT2G29110 |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
7.086 |
0.217 |
| 113 |
AT5G01550 |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
7.078 |
0.299 |
| 114 |
AT4G08770 |
AT4G08770
|
Prx37, peroxidase 37 |
7.070 |
0.792 |
| 115 |
AT1G13520 |
AT1G13520
|
unknown |
7.050 |
no data |
| 116 |
AT1G65690 |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
7.038 |
1.024 |
| 117 |
AT3G48650 |
AT3G48650
|
[pseudogene] |
7.024 |
0.612 |
| 118 |
AT5G25260 |
AT5G25260
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
7.016 |
no data |
| 119 |
AT3G29000 |
AT3G29000
|
[Calcium-binding EF-hand family protein] |
7.007 |
no data |
| 120 |
AT2G23270 |
AT2G23270
|
unknown |
7.004 |
0.172 |
| 121 |
AT4G23150 |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
6.955 |
0.057 |
| 122 |
AT1G30720 |
AT1G30720
|
[FAD-binding Berberine family protein] |
6.924 |
no data |
| 123 |
AT4G16820 |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
6.924 |
0.090 |
| 124 |
AT2G02010 |
AT2G02010
|
GAD4, glutamate decarboxylase 4 |
6.921 |
no data |
| 125 |
AT5G25250 |
AT5G25250
|
FLOT1, flotillin 1 |
6.920 |
no data |
| 126 |
AT2G39530 |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
6.913 |
0.562 |
| 127 |
AT4G23210 |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
6.903 |
0.758 |
| 128 |
AT2G39518 |
AT2G39518
|
[Uncharacterised protein family (UPF0497)] |
6.865 |
no data |
| 129 |
AT5G22520 |
AT5G22520
|
unknown |
6.851 |
no data |
| 130 |
AT1G32960 |
AT1G32960
|
ATSBT3.3, SBT3.3 |
6.849 |
0.689 |
| 131 |
AT5G11140 |
AT5G11140
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
6.840 |
no data |
| 132 |
AT3G13433 |
AT3G13433
|
unknown |
6.831 |
no data |
| 133 |
AT4G08555 |
AT4G08555
|
unknown |
6.821 |
no data |
| 134 |
AT5G01380 |
AT5G01380
|
[Homeodomain-like superfamily protein] |
6.801 |
0.213 |
| 135 |
AT5G02490 |
AT5G02490
|
AtHsp70-2, Hsp70-2 |
6.796 |
-0.111 |
| 136 |
AT5G57480 |
AT5G57480
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
6.795 |
0.317 |
| 137 |
AT1G27020 |
AT1G27020
|
unknown |
6.782 |
no data |
| 138 |
AT1G36620 |
AT1G36620
|
[copia-like retrotransposon family, has a 3.6e-54 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
6.757 |
-0.028 |
| 139 |
AT4G03450 |
AT4G03450
|
[Ankyrin repeat family protein] |
6.741 |
0.134 |
| 140 |
AT3G15356 |
AT3G15356
|
[Legume lectin family protein] |
6.739 |
no data |
| 141 |
AT1G68765 |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
6.730 |
0.213 |
| 142 |
AT2G22880 |
AT2G22880
|
[VQ motif-containing protein] |
6.722 |
1.092 |
| 143 |
AT5G36970 |
AT5G36970
|
NHL25, NDR1/HIN1-like 25 |
6.713 |
no data |
| 144 |
AT2G25735 |
AT2G25735
|
unknown |
6.702 |
0.857 |
| 145 |
AT4G21390 |
AT4G21390
|
B120 |
6.691 |
0.882 |
| 146 |
AT3G25490 |
AT3G25490
|
[Protein kinase family protein] |
6.691 |
0.008 |
| 147 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
6.691 |
1.010 |
| 148 |
AT2G15490 |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
6.684 |
0.417 |
| 149 |
AT5G52760 |
AT5G52760
|
[Copper transport protein family] |
6.670 |
0.545 |
| 150 |
AT1G56240 |
AT1G56240
|
AtPP2-B13, phloem protein 2-B13, PP2-B13, phloem protein 2-B13 |
6.670 |
no data |
| 151 |
AT3G04220 |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
6.665 |
0.264 |
| 152 |
AT4G23190 |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
6.665 |
0.672 |
| 153 |
AT1G57650 |
AT1G57650
|
[ATP binding] |
6.649 |
0.114 |
| 154 |
AT1G61800 |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
6.646 |
0.487 |
| 155 |
AT1G26390 |
AT1G26390
|
[FAD-binding Berberine family protein] |
6.642 |
0.636 |
| 156 |
AT3G49620 |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
6.630 |
1.524 |
| 157 |
AT1G35210 |
AT1G35210
|
unknown |
6.618 |
0.313 |
| 158 |
AT3G25250 |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
6.617 |
0.406 |
| 159 |
AT1G29715 |
AT1G29715
|
[pseudogene] |
6.614 |
no data |
| 160 |
AT1G17147 |
AT1G17147
|
[VQ motif-containing protein] |
6.600 |
no data |
| 161 |
AT1G44130 |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
6.580 |
0.122 |
| 162 |
AT3G55840 |
AT3G55840
|
[Hs1pro-1 protein] |
6.575 |
0.559 |
| 163 |
AT1G05880 |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
6.572 |
-0.039 |
| 164 |
AT3G28210 |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
6.571 |
0.839 |
| 165 |
AT4G02380 |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
6.570 |
0.505 |
| 166 |
AT1G57630 |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
6.561 |
0.783 |
| 167 |
AT4G14365 |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
6.559 |
0.395 |
| 168 |
AT1G73810 |
AT1G73810
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
6.554 |
no data |
| 169 |
AT2G34500 |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
6.547 |
0.496 |
| 170 |
AT3G48850 |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
6.535 |
0.586 |
| 171 |
AT3G01290 |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
6.519 |
0.267 |
| 172 |
AT1G26410 |
AT1G26410
|
[FAD-binding Berberine family protein] |
6.517 |
0.505 |
| 173 |
AT1G07135 |
AT1G07135
|
[glycine-rich protein] |
6.516 |
no data |
| 174 |
AT4G24110 |
AT4G24110
|
unknown |
6.513 |
0.395 |
| 175 |
AT1G35230 |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
6.509 |
0.674 |
| 176 |
AT1G51850 |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
6.508 |
0.173 |
| 177 |
AT4G04510 |
AT4G04510
|
CRK38, cysteine-rich RLK (RECEPTOR-like protein kinase) 38 |
6.484 |
0.039 |
| 178 |
AT3G50930 |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
6.451 |
0.655 |
| 179 |
AT1G36610 |
AT1G36610
|
[gypsy-like retrotransposon family, has a 9.1e-283 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
6.448 |
no data |
| 180 |
AT2G47140 |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
6.445 |
0.315 |
| 181 |
AT1G02930 |
AT1G02930
|
ATGST1, ARABIDOPSIS GLUTATHIONE S-TRANSFERASE 1, ATGSTF3, ARABIDOPSIS THALIANA GLUATIONE S-TRANSFERASE F3, ATGSTF6, GLUTATHIONE S-TRANSFERASE, ERD11, EARLY RESPONSIVE TO DEHYDRATION 11, GST1, GLUTATHIONE S-TRANSFERASE 1, GSTF6, glutathione S-transferase 6 |
6.440 |
no data |
| 182 |
AT3G63380 |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
6.439 |
0.676 |
| 183 |
AT4G21840 |
AT4G21840
|
ATMSRB8, methionine sulfoxide reductase B8, MSRB8, methionine sulfoxide reductase B8 |
6.439 |
no data |
| 184 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
6.433 |
0.667 |
| 185 |
AT4G25070 |
AT4G25070
|
unknown |
6.427 |
0.155 |
| 186 |
AT4G22030 |
AT4G22030
|
unknown |
6.421 |
0.012 |
| 187 |
AT1G53620 |
AT1G53620
|
unknown |
6.417 |
no data |
| 188 |
AT4G03460 |
AT4G03460
|
[Ankyrin repeat family protein] |
6.409 |
no data |
| 189 |
AT5G52640 |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
6.391 |
-0.312 |
| 190 |
AT1G21100 |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
6.391 |
0.931 |
| 191 |
AT4G25110 |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
6.378 |
0.137 |
| 192 |
AT3G01760 |
AT3G01760
|
[Transmembrane amino acid transporter family protein] |
6.365 |
no data |
| 193 |
AT5G38900 |
AT5G38900
|
[Thioredoxin superfamily protein] |
6.361 |
0.634 |
| 194 |
AT2G17740 |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
6.360 |
0.623 |
| 195 |
AT5G27420 |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
6.358 |
0.701 |
| 196 |
AT3G15518 |
AT3G15518
|
unknown |
6.352 |
no data |
| 197 |
AT1G21240 |
AT1G21240
|
WAK3, wall associated kinase 3 |
6.352 |
0.199 |
| 198 |
AT3G18250 |
AT3G18250
|
[Putative membrane lipoprotein] |
6.344 |
0.515 |
| 199 |
AT3G44830 |
AT3G44830
|
[Lecithin:cholesterol acyltransferase family protein] |
6.342 |
-0.008 |
| 200 |
AT2G32130 |
AT2G32130
|
unknown |
6.333 |
no data |
| 201 |
AT4G15975 |
AT4G15975
|
[RING/U-box superfamily protein] |
6.325 |
no data |
| 202 |
AT4G38560 |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
6.309 |
0.268 |
| 203 |
AT4G35180 |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
6.305 |
0.371 |
| 204 |
AT1G07160 |
AT1G07160
|
[Protein phosphatase 2C family protein] |
6.302 |
0.024 |
| 205 |
AT5G22380 |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
6.281 |
0.642 |
| 206 |
AT5G51060 |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
6.278 |
0.135 |
| 207 |
AT3G50140 |
AT3G50140
|
unknown |
6.278 |
0.005 |
| 208 |
AT4G39610 |
AT4G39610
|
unknown |
6.272 |
-0.064 |
| 209 |
AT2G38470 |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
6.272 |
0.458 |
| 210 |
AT1G43000 |
AT1G43000
|
[PLATZ transcription factor family protein] |
6.258 |
0.068 |
| 211 |
AT2G26150 |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
6.246 |
-0.371 |
| 212 |
AT3G13950 |
AT3G13950
|
unknown |
6.240 |
0.665 |
| 213 |
AT5G25930 |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
6.236 |
0.622 |
| 214 |
AT5G45090 |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
6.214 |
0.064 |
| 215 |
AT5G52400 |
AT5G52400
|
CYP715A1, cytochrome P450, family 715, subfamily A, polypeptide 1 |
6.209 |
0.571 |
| 216 |
AT4G18250 |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
6.209 |
0.503 |
| 217 |
AT2G32190 |
AT2G32190
|
unknown |
6.186 |
no data |
| 218 |
AT2G32140 |
AT2G32140
|
[transmembrane receptors] |
6.183 |
0.929 |
| 219 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
6.182 |
no data |
| 220 |
AT2G32030 |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
6.179 |
0.475 |
| 221 |
AT3G25610 |
AT3G25610
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
6.171 |
0.331 |
| 222 |
AT1G59865 |
AT1G59865
|
unknown |
6.157 |
no data |
| 223 |
AT1G56250 |
AT1G56250
|
AtPP2-B14, phloem protein 2-B14, PP2-B14, phloem protein 2-B14, VBF, VIP1-binding F-box protein |
6.157 |
no data |
| 224 |
AT2G37980 |
AT2G37980
|
[O-fucosyltransferase family protein] |
6.155 |
0.344 |
| 225 |
AT1G19250 |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
6.131 |
0.141 |
| 226 |
AT2G27660 |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
6.122 |
0.294 |
| 227 |
AT3G11340 |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
6.121 |
0.452 |
| 228 |
AT5G49520 |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
6.118 |
0.920 |
| 229 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
6.117 |
0.507 |
| 230 |
AT2G36690 |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
6.109 |
0.325 |
| 231 |
AT5G67450 |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
6.107 |
0.155 |
| 232 |
AT4G17490 |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
6.099 |
0.489 |
| 233 |
AT5G67340 |
AT5G67340
|
[ARM repeat superfamily protein] |
6.099 |
0.622 |
| 234 |
AT2G45760 |
AT2G45760
|
BAL, BON ASSOCIATION PROTEIN 1-LIKE, BAP2, BON association protein 2 |
6.099 |
no data |
| 235 |
AT1G65390 |
AT1G65390
|
ATPP2-A5, phloem protein 2 A5, PP2-A5, phloem protein 2 A5 |
6.098 |
0.726 |
| 236 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
6.087 |
0.597 |
| 237 |
AT5G01100 |
AT5G01100
|
FRB1, FRIABLE 1 |
6.085 |
no data |
| 238 |
AT1G21326 |
AT1G21326
|
[VQ motif-containing protein] |
6.083 |
no data |
| 239 |
AT1G51790 |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
6.082 |
0.351 |
| 240 |
AT5G25920 |
AT5G25920
|
unknown |
6.082 |
0.003 |
| 241 |
AT2G04495 |
AT2G04495
|
unknown |
6.082 |
no data |
| 242 |
AT1G33730 |
AT1G33730
|
CYP76C5, cytochrome P450, family 76, subfamily C, polypeptide 5 |
6.058 |
no data |
| 243 |
AT4G40020 |
AT4G40020
|
[Myosin heavy chain-related protein] |
6.045 |
0.090 |
| 244 |
AT2G13810 |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
6.044 |
0.165 |
| 245 |
AT4G04490 |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
6.044 |
0.298 |
| 246 |
AT1G51913 |
AT1G51913
|
[LOCATED IN: endomembrane system] |
6.031 |
no data |
| 247 |
AT3G45330 |
AT3G45330
|
LecRK-I.1, L-type lectin receptor kinase I.1 |
6.027 |
-0.011 |
| 248 |
AT4G30280 |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
6.025 |
0.697 |
| 249 |
AT4G17660 |
AT4G17660
|
[Protein kinase superfamily protein] |
6.021 |
0.076 |
| 250 |
AT4G11370 |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
6.015 |
0.280 |
| 251 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
6.002 |
0.876 |
| 252 |
AT2G14290 |
AT2G14290
|
unknown |
5.996 |
0.766 |
| 253 |
AT2G29460 |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
5.995 |
1.085 |
| 254 |
AT2G24850 |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
5.987 |
0.651 |
| 255 |
AT5G62480 |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
5.985 |
0.287 |
| 256 |
AT2G45220 |
AT2G45220
|
PME17, pectin methylesterase 17 |
5.982 |
0.675 |
| 257 |
AT1G05575 |
AT1G05575
|
unknown |
5.970 |
1.162 |
| 258 |
AT1G62300 |
AT1G62300
|
ATWRKY6, WRKY6 |
5.965 |
0.601 |
| 259 |
AT4G26200 |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
5.964 |
0.117 |
| 260 |
AT1G09932 |
AT1G09932
|
[Phosphoglycerate mutase family protein] |
5.960 |
no data |
| 261 |
AT5G51440 |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
5.949 |
-0.687 |
| 262 |
AT2G32210 |
AT2G32210
|
unknown |
5.938 |
no data |
| 263 |
AT4G09300 |
AT4G09300
|
[LisH and RanBPM domains containing protein] |
5.922 |
0.028 |
| 264 |
AT2G04070 |
AT2G04070
|
[MATE efflux family protein] |
5.916 |
0.083 |
| 265 |
AT3G53150 |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
5.916 |
0.182 |
| 266 |
AT1G10040 |
AT1G10040
|
[alpha/beta-Hydrolases superfamily protein] |
5.911 |
no data |
| 267 |
AT3G22160 |
AT3G22160
|
JAV1, jasmonate-associated VQ motif gene 1 |
5.906 |
0.510 |
| 268 |
AT1G11100 |
AT1G11100
|
[SNF2 domain-containing protein / helicase domain-containing protein / zinc finger protein-related] |
5.904 |
-0.024 |
| 269 |
AT3G62150 |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
5.891 |
0.399 |
| 270 |
AT2G46400 |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
5.890 |
0.401 |
| 271 |
AT2G15480 |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
5.890 |
0.596 |
| 272 |
AT4G23220 |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
5.869 |
0.715 |
| 273 |
AT5G66640 |
AT5G66640
|
DAR3, DA1-related protein 3 |
5.868 |
0.500 |
| 274 |
AT1G65484 |
AT1G65484
|
unknown |
5.867 |
no data |
| 275 |
AT4G37370 |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
5.860 |
0.701 |
| 276 |
AT1G18390 |
AT1G18390
|
[Protein kinase superfamily protein] |
5.859 |
0.541 |
| 277 |
AT2G29720 |
AT2G29720
|
CTF2B |
5.854 |
no data |
| 278 |
AT1G19180 |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
5.839 |
0.900 |
| 279 |
AT5G38310 |
AT5G38310
|
unknown |
5.826 |
0.227 |
| 280 |
AT2G25440 |
AT2G25440
|
AtRLP20, receptor like protein 20, RLP20, receptor like protein 20 |
5.824 |
0.323 |
| 281 |
AT2G42350 |
AT2G42350
|
[RING/U-box superfamily protein] |
5.821 |
0.356 |
| 282 |
AT1G74360 |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
5.820 |
0.677 |
| 283 |
AT3G16530 |
AT3G16530
|
[Legume lectin family protein] |
5.819 |
0.406 |
| 284 |
AT2G41100 |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
5.813 |
0.224 |
| 285 |
AT1G51620 |
AT1G51620
|
[Protein kinase superfamily protein] |
5.813 |
0.394 |
| 286 |
AT2G40095 |
AT2G40095
|
[Alpha/beta hydrolase related protein] |
5.812 |
no data |
| 287 |
AT5G54490 |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
5.811 |
0.597 |
| 288 |
AT5G26340 |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
5.810 |
0.841 |
| 289 |
AT4G11480 |
AT4G11480
|
CRK32, cysteine-rich RLK (RECEPTOR-like protein kinase) 32 |
5.804 |
0.121 |
| 290 |
AT5G18470 |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
5.801 |
0.748 |
| 291 |
AT5G64120 |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
5.800 |
0.444 |
| 292 |
AT2G43000 |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
5.790 |
0.329 |
| 293 |
AT1G24140 |
AT1G24140
|
[Matrixin family protein] |
5.788 |
0.543 |
| 294 |
AT1G27730 |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
5.785 |
0.833 |
| 295 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
5.777 |
1.263 |
| 296 |
AT1G65481 |
AT1G65481
|
unknown |
5.773 |
no data |
| 297 |
AT5G67310 |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
5.760 |
0.605 |
| 298 |
AT5G47230 |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
5.759 |
0.254 |
| 299 |
AT1G13340 |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
5.753 |
0.621 |
| 300 |
AT1G01480 |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
5.749 |
0.563 |
| 301 |
AT3G24750 |
AT3G24750
|
unknown |
-4.575 |
-0.081 |
| 302 |
AT1G70270 |
AT1G70270
|
unknown |
-4.143 |
-0.267 |
| 303 |
AT3G22100 |
AT3G22100
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-3.815 |
no data |
| 304 |
AT2G22810 |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-3.802 |
-0.156 |
| 305 |
AT5G40790 |
AT5G40790
|
unknown |
-3.624 |
no data |
| 306 |
AT1G29460 |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-3.578 |
-0.498 |
| 307 |
AT5G13170 |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-3.538 |
-0.281 |
| 308 |
AT1G78440 |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-3.519 |
-0.066 |
| 309 |
AT4G25420 |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-3.489 |
-0.500 |
| 310 |
AT1G51090 |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-3.449 |
0.716 |
| 311 |
AT5G18010 |
AT5G18010
|
SAUR19, small auxin up RNA 19 |
-3.408 |
no data |
| 312 |
AT3G03840 |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-3.392 |
-0.416 |
| 313 |
AT2G40610 |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-3.389 |
-0.584 |
| 314 |
AT5G18080 |
AT5G18080
|
SAUR24, small auxin up RNA 24 |
-3.358 |
no data |
| 315 |
AT4G33790 |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-3.346 |
-0.133 |
| 316 |
AT4G06534 |
AT4G06534
|
unknown |
-3.238 |
no data |
| 317 |
AT1G29450 |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-3.237 |
-0.351 |
| 318 |
AT5G18050 |
AT5G18050
|
SAUR22, SMALL AUXIN UP RNA 22 |
-3.225 |
no data |
| 319 |
AT1G29420 |
AT1G29420
|
SAUR61, SMALL AUXIN UPREGULATED RNA 61 |
-3.219 |
no data |
| 320 |
AT5G15500 |
AT5G15500
|
[Ankyrin repeat family protein] |
-3.156 |
no data |
| 321 |
AT5G18020 |
AT5G18020
|
SAUR20, SMALL AUXIN UP RNA 20 |
-3.068 |
no data |
| 322 |
AT3G62610 |
AT3G62610
|
ATMYB11, myb domain protein 11, MYB11, myb domain protein 11, PFG2, PRODUCTION OF FLAVONOL GLYCOSIDES 2 |
-3.042 |
-0.084 |
| 323 |
AT1G60190 |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-3.039 |
0.606 |
| 324 |
AT5G39860 |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-3.030 |
-0.031 |
| 325 |
AT5G24880 |
AT5G24880
|
unknown |
-2.974 |
-0.046 |
| 326 |
AT5G18030 |
AT5G18030
|
[SAUR-like auxin-responsive protein family ] |
-2.968 |
no data |
| 327 |
AT1G29500 |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-2.962 |
-0.508 |
| 328 |
AT3G42800 |
AT3G42800
|
unknown |
-2.958 |
-0.176 |
| 329 |
AT3G03830 |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-2.885 |
-0.174 |
| 330 |
AT1G12672 |
AT1G12672
|
unknown |
-2.866 |
no data |
| 331 |
AT4G32280 |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-2.866 |
0.128 |
| 332 |
AT4G38850 |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-2.843 |
-0.289 |
| 333 |
AT1G07430 |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-2.823 |
0.942 |
| 334 |
AT1G15580 |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-2.775 |
0.062 |
| 335 |
AT3G21330 |
AT3G21330
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-2.762 |
no data |
| 336 |
AT4G16590 |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
-2.753 |
0.727 |
| 337 |
AT1G29430 |
AT1G29430
|
SAUR62, SMALL AUXIN UPREGULATED RNA 62 |
-2.727 |
no data |
| 338 |
AT5G02760 |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-2.707 |
-0.712 |
| 339 |
AT1G20190 |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-2.697 |
-0.446 |
| 340 |
AT1G29510 |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-2.657 |
-0.736 |
| 341 |
AT1G29440 |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-2.652 |
-0.720 |
| 342 |
AT1G69140 |
AT1G69140
|
[pseudogene] |
-2.649 |
no data |
| 343 |
AT3G03820 |
AT3G03820
|
SAUR29, SMALL AUXIN UP RNA 29 |
-2.645 |
no data |
| 344 |
AT5G54585 |
AT5G54585
|
unknown |
-2.643 |
no data |
| 345 |
AT1G56600 |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-2.615 |
0.288 |
| 346 |
AT5G03120 |
AT5G03120
|
unknown |
-2.580 |
-0.598 |
| 347 |
AT4G04840 |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-2.570 |
0.155 |
| 348 |
AT4G14819 |
AT4G14819
|
unknown |
-2.569 |
no data |
| 349 |
AT4G14560 |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-2.569 |
-0.208 |
| 350 |
AT5G66590 |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-2.566 |
0.139 |
| 351 |
AT4G01080 |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-2.562 |
-0.030 |
| 352 |
AT5G34810 |
AT5G34810
|
[pseudogene] |
-2.557 |
no data |
| 353 |
AT5G05430 |
AT5G05430
|
[RNA-binding protein] |
-2.547 |
0.031 |
| 354 |
AT1G26945 |
AT1G26945
|
KDR, KIDARI, PRE6, PACLOBUTRAZOL RESISTANCE 6 |
-2.540 |
no data |
| 355 |
AT2G07140 |
AT2G07140
|
[F-box and associated interaction domains-containing protein] |
-2.538 |
no data |
| 356 |
AT3G24518 |
AT3G24518
|
other RNA |
-2.531 |
no data |
| 357 |
AT1G62510 |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-2.526 |
-0.068 |
| 358 |
AT2G26710 |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-2.520 |
-0.689 |
| 359 |
AT2G30300 |
AT2G30300
|
[Major facilitator superfamily protein] |
-2.515 |
-0.055 |
| 360 |
AT3G03850 |
AT3G03850
|
SAUR26, SMALL AUXIN UP RNA 26 |
-2.510 |
no data |
| 361 |
AT5G59310 |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-2.508 |
0.287 |
| 362 |
AT2G16367 |
AT2G16367
|
|
-2.502 |
no data |
| 363 |
AT4G13800 |
AT4G13800
|
unknown |
-2.502 |
0.221 |
| 364 |
AT3G08860 |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-2.489 |
0.213 |
| 365 |
AT1G29660 |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-2.488 |
-0.414 |
| 366 |
AT3G27250 |
AT3G27250
|
unknown |
-2.488 |
no data |
| 367 |
AT4G04810 |
AT4G04810
|
ATMSRB4, methionine sulfoxide reductase B4, MSRB4, methionine sulfoxide reductase B4 |
-2.467 |
no data |
| 368 |
AT5G66580 |
AT5G66580
|
unknown |
-2.467 |
no data |
| 369 |
AT3G62070 |
AT3G62070
|
unknown |
-2.458 |
no data |
| 370 |
AT1G06980 |
AT1G06980
|
unknown |
-2.458 |
-0.405 |
| 371 |
AT5G18060 |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-2.456 |
-0.365 |
| 372 |
AT5G19470 |
AT5G19470
|
atnudt24, nudix hydrolase homolog 24, NUDT24, nudix hydrolase homolog 24, NUDX24, nudix hydrolase homolog 24 |
-2.446 |
no data |
| 373 |
AT5G10250 |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-2.434 |
-0.511 |
| 374 |
AT4G08570 |
AT4G08570
|
[Heavy metal transport/detoxification superfamily protein ] |
-2.428 |
no data |
| 375 |
AT1G52830 |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-2.428 |
0.189 |
| 376 |
AT5G24155 |
AT5G24155
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-2.404 |
no data |
| 377 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-2.401 |
0.837 |
| 378 |
AT5G25390 |
AT5G25390
|
SHN3, shine3 |
-2.395 |
0.047 |
| 379 |
AT1G16400 |
AT1G16400
|
CYP79F2, cytochrome P450, family 79, subfamily F, polypeptide 2 |
-2.392 |
no data |
| 380 |
AT2G15060 |
AT2G15060
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 1.8e-63 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
-2.387 |
0.051 |
| 381 |
AT1G22370 |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
-2.386 |
-0.048 |
| 382 |
AT1G57590 |
AT1G57590
|
[Pectinacetylesterase family protein] |
-2.381 |
0.260 |
| 383 |
AT5G05840 |
AT5G05840
|
unknown |
-2.376 |
0.052 |
| 384 |
AT5G65730 |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-2.369 |
-0.527 |
| 385 |
AT5G01790 |
AT5G01790
|
unknown |
-2.367 |
-0.294 |
| 386 |
AT4G14550 |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-2.356 |
0.113 |
| 387 |
AT2G42380 |
AT2G42380
|
ATBZIP34, BZIP34 |
-2.355 |
-0.393 |
| 388 |
AT1G19960 |
AT1G19960
|
unknown |
-2.355 |
-0.237 |
| 389 |
AT5G52900 |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-2.352 |
-0.097 |
| 390 |
AT3G58070 |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-2.351 |
-0.537 |
| 391 |
AT1G04240 |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-2.351 |
-0.321 |
| 392 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-2.350 |
0.726 |
| 393 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-2.350 |
0.615 |
| 394 |
AT1G07490 |
AT1G07490
|
DVL9, DEVIL 9, RTFL3, ROTUNDIFOLIA like 3 |
-2.336 |
no data |
| 395 |
AT3G58120 |
AT3G58120
|
ATBZIP61, BZIP61 |
-2.334 |
-0.767 |
| 396 |
AT2G44230 |
AT2G44230
|
unknown |
-2.333 |
0.255 |
| 397 |
AT3G24460 |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-2.326 |
-0.579 |
| 398 |
AT4G18960 |
AT4G18960
|
AG, AGAMOUS |
-2.324 |
-0.021 |
| 399 |
AT4G25433 |
AT4G25433
|
[peptidoglycan-binding LysM domain-containing protein] |
-2.318 |
no data |
| 400 |
AT5G62280 |
AT5G62280
|
unknown |
-2.308 |
-0.400 |
| 401 |
AT2G40440 |
AT2G40440
|
[BTB/POZ domain-containing protein] |
-2.307 |
-0.044 |
| 402 |
AT5G47370 |
AT5G47370
|
HAT2 |
-2.306 |
-0.278 |
| 403 |
AT3G10185 |
AT3G10185
|
[Gibberellin-regulated family protein] |
-2.290 |
no data |
| 404 |
AT5G64770 |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-2.285 |
-0.142 |
| 405 |
AT2G32100 |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-2.265 |
-0.342 |
| 406 |
AT5G10990 |
AT5G10990
|
SAUR69, SMALL AUXIN UPREGULATED RNA 69 |
-2.263 |
no data |
| 407 |
AT3G26960 |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-2.251 |
-0.564 |
| 408 |
AT2G22930 |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-2.248 |
0.265 |
| 409 |
AT5G41400 |
AT5G41400
|
[RING/U-box superfamily protein] |
-2.240 |
-0.119 |
| 410 |
AT1G16410 |
AT1G16410
|
BUS1, BUSHY 1, CYP79F1, cytochrome p450 79f1, SPS1, SUPERSHOOT 1 |
-2.240 |
no data |
| 411 |
AT5G20110 |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-2.234 |
-0.403 |
| 412 |
AT2G39800 |
AT2G39800
|
ATP5CS, P5CS1, delta1-pyrroline-5-carboxylate synthase 1 |
-2.210 |
no data |
| 413 |
AT1G29110 |
AT1G29110
|
[Cysteine proteinases superfamily protein] |
-2.202 |
-0.077 |
| 414 |
AT1G01250 |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-2.189 |
-0.203 |
| 415 |
AT2G47780 |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-2.183 |
0.253 |
| 416 |
AT2G41190 |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-2.180 |
0.167 |
| 417 |
AT1G52750 |
AT1G52750
|
[alpha/beta-Hydrolases superfamily protein] |
-2.179 |
no data |
| 418 |
AT1G74670 |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-2.174 |
-0.422 |
| 419 |
AT2G05995 |
AT2G05995
|
other RNA |
-2.168 |
no data |
| 420 |
AT2G22200 |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-2.168 |
0.744 |
| 421 |
AT3G25717 |
AT3G25717
|
DVL6, DEVIL 6, RTFL16, ROTUNDIFOLIA like 16 |
-2.168 |
no data |
| 422 |
AT1G09350 |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-2.159 |
0.576 |
| 423 |
AT4G26790 |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-2.154 |
-0.586 |
| 424 |
AT4G06536 |
AT4G06536
|
[SPla/RYanodine receptor (SPRY) domain-containing protein] |
-2.132 |
no data |
| 425 |
AT3G56275 |
AT3G56275
|
[expressed protein] |
-2.131 |
no data |
| 426 |
AT5G55620 |
AT5G55620
|
unknown |
-2.128 |
-0.418 |
| 427 |
AT5G07700 |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
-2.127 |
0.033 |
| 428 |
AT4G35320 |
AT4G35320
|
unknown |
-2.114 |
-0.252 |
| 429 |
AT5G41780 |
AT5G41780
|
[myosin heavy chain-related] |
-2.113 |
-0.080 |
| 430 |
AT1G71140 |
AT1G71140
|
[MATE efflux family protein] |
-2.106 |
0.149 |
| 431 |
AT4G04830 |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-2.101 |
0.085 |
| 432 |
AT3G23870 |
AT3G23870
|
unknown |
-2.090 |
0.227 |
| 433 |
AT3G25670 |
AT3G25670
|
[Leucine-rich repeat (LRR) family protein] |
-2.089 |
-0.259 |
| 434 |
AT4G17770 |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-2.084 |
-0.466 |
| 435 |
AT1G76955 |
AT1G76955
|
[Expressed protein] |
-2.084 |
no data |
| 436 |
AT1G26210 |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-2.080 |
-0.483 |
| 437 |
AT1G12090 |
AT1G12090
|
ELP, extensin-like protein |
-2.079 |
-0.088 |
| 438 |
AT5G01880 |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-2.069 |
-0.168 |
| 439 |
AT3G10150 |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-2.069 |
-0.461 |
| 440 |
AT4G10150 |
AT4G10150
|
[RING/U-box superfamily protein] |
-2.067 |
no data |
| 441 |
AT5G51790 |
AT5G51790
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-2.065 |
no data |
| 442 |
AT3G22740 |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-2.061 |
0.427 |
| 443 |
AT4G34550 |
AT4G34550
|
unknown |
-2.061 |
no data |
| 444 |
AT1G22380 |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
-2.051 |
0.055 |
| 445 |
AT3G56080 |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-2.045 |
-0.047 |
| 446 |
AT3G24520 |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-2.039 |
0.467 |
| 447 |
AT1G64780 |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-2.037 |
-0.559 |
| 448 |
AT3G44990 |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-2.035 |
-0.738 |
| 449 |
AT2G29710 |
AT2G29710
|
[UDP-Glycosyltransferase superfamily protein] |
-2.032 |
no data |
| 450 |
AT1G73700 |
AT1G73700
|
[MATE efflux family protein] |
-2.032 |
0.071 |
| 451 |
AT5G63160 |
AT5G63160
|
BT1, BTB and TAZ domain protein 1 |
-2.028 |
no data |
| 452 |
AT4G19170 |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-2.027 |
-0.114 |
| 453 |
AT5G52930 |
AT5G52930
|
unknown |
-2.024 |
no data |
| 454 |
AT5G15310 |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-2.022 |
-0.352 |
| 455 |
AT2G01505 |
AT2G01505
|
CLE16, CLAVATA3/ESR-RELATED 16 |
-2.018 |
no data |
| 456 |
AT3G22560 |
AT3G22560
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-2.011 |
no data |
| 457 |
AT5G52940 |
AT5G52940
|
unknown |
-2.008 |
no data |
| 458 |
AT2G46660 |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
-2.003 |
-0.287 |
| 459 |
AT2G27420 |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-2.001 |
-1.219 |
| 460 |
AT1G79170 |
AT1G79170
|
unknown |
-1.997 |
-0.091 |
| 461 |
AT5G43870 |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-1.996 |
-0.460 |
| 462 |
AT3G61880 |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
-1.994 |
-0.052 |
| 463 |
AT1G56650 |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-1.986 |
0.749 |
| 464 |
AT3G50840 |
AT3G50840
|
[Phototropic-responsive NPH3 family protein] |
-1.983 |
no data |
| 465 |
AT2G15970 |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
-1.982 |
0.124 |
| 466 |
AT5G50800 |
AT5G50800
|
AtSWEET13, SWEET13 |
-1.982 |
-0.281 |
| 467 |
AT3G30460 |
AT3G30460
|
[RING/U-box superfamily protein] |
-1.978 |
-0.244 |
| 468 |
AT2G32560 |
AT2G32560
|
[F-box family protein] |
-1.969 |
-0.064 |
| 469 |
AT5G06760 |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-1.968 |
0.487 |
| 470 |
AT4G38770 |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-1.963 |
-0.189 |
| 471 |
AT1G67900 |
AT1G67900
|
[Phototropic-responsive NPH3 family protein] |
-1.955 |
no data |
| 472 |
AT2G38750 |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-1.952 |
0.422 |
| 473 |
AT1G26208 |
AT1G26208
|
other RNA |
-1.952 |
no data |
| 474 |
AT4G29905 |
AT4G29905
|
unknown |
-1.951 |
no data |
| 475 |
AT4G04760 |
AT4G04760
|
[Major facilitator superfamily protein] |
-1.949 |
-0.047 |
| 476 |
AT4G09920 |
AT4G09920
|
[FBD, F-box and Leucine Rich Repeat domains containing protein] |
-1.923 |
no data |
| 477 |
AT2G35950 |
AT2G35950
|
EDA12, embryo sac development arrest 12 |
-1.922 |
-0.040 |
| 478 |
AT2G15020 |
AT2G15020
|
unknown |
-1.922 |
-0.571 |
| 479 |
AT3G51895 |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-1.921 |
-0.058 |
| 480 |
AT3G05936 |
AT3G05936
|
unknown |
-1.920 |
no data |
| 481 |
AT4G23990 |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
-1.918 |
-0.149 |
| 482 |
AT3G58990 |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-1.916 |
-0.147 |
| 483 |
AT1G02820 |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-1.914 |
-0.121 |
| 484 |
AT5G38100 |
AT5G38100
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.913 |
no data |
| 485 |
AT5G44005 |
AT5G44005
|
unknown |
-1.909 |
no data |
| 486 |
AT3G28080 |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-1.902 |
-0.451 |
| 487 |
AT3G17640 |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-1.900 |
-0.488 |
| 488 |
AT1G01520 |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-1.892 |
-0.678 |
| 489 |
AT2G40130 |
AT2G40130
|
SMXL8, SMAX1-like 8 |
-1.884 |
-0.062 |
| 490 |
AT5G45950 |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.881 |
-0.085 |
| 491 |
AT2G40475 |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-1.880 |
0.046 |
| 492 |
AT1G60590 |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.877 |
-0.918 |
| 493 |
AT4G26530 |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-1.877 |
-0.142 |
| 494 |
AT5G42040 |
AT5G42040
|
RPN12b, regulatory particle non-ATPase 12B |
-1.874 |
no data |
| 495 |
AT1G68870 |
AT1G68870
|
ATSOFL2, SOB FIVE-LIKE 2, SOFL2, SOB five-like 2 |
-1.872 |
no data |
| 496 |
AT5G02890 |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-1.862 |
-0.121 |
| 497 |
AT3G02020 |
AT3G02020
|
AK3, aspartate kinase 3 |
-1.858 |
no data |
| 498 |
AT3G46650 |
AT3G46650
|
[UDP-Glycosyltransferase superfamily protein] |
-1.858 |
no data |
| 499 |
AT2G39920 |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-1.855 |
0.312 |
| 500 |
AT4G03070 |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-1.854 |
-0.517 |
| 501 |
AT1G15260 |
AT1G15260
|
unknown |
-1.854 |
-0.442 |
| 502 |
AT5G45830 |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
-1.853 |
0.068 |
| 503 |
AT2G41312 |
AT2G41312
|
other RNA |
-1.852 |
no data |
| 504 |
AT1G20470 |
AT1G20470
|
SAUR60, SMALL AUXIN UPREGULATED RNA 60 |
-1.849 |
no data |
| 505 |
AT1G68600 |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-1.848 |
0.285 |
| 506 |
AT5G41900 |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-1.842 |
0.047 |
| 507 |
AT3G21670 |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-1.840 |
0.005 |
| 508 |
AT4G02630 |
AT4G02630
|
[Protein kinase superfamily protein] |
-1.835 |
-0.389 |
| 509 |
AT2G23130 |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-1.831 |
-0.619 |
| 510 |
AT1G78230 |
AT1G78230
|
[Outer arm dynein light chain 1 protein] |
-1.829 |
no data |
| 511 |
AT4G09890 |
AT4G09890
|
unknown |
-1.827 |
-0.490 |
| 512 |
AT5G49740 |
AT5G49740
|
ATFRO7, FERRIC REDUCTION OXIDASE 7, FRO7, ferric reduction oxidase 7 |
-1.820 |
no data |
| 513 |
AT4G17340 |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-1.820 |
0.103 |
| 514 |
AT2G40435 |
AT2G40435
|
unknown |
-1.819 |
0.488 |
| 515 |
AT1G48100 |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-1.811 |
-0.206 |
| 516 |
AT1G66100 |
AT1G66100
|
[Plant thionin] |
-1.810 |
0.238 |
| 517 |
AT3G44900 |
AT3G44900
|
ATCHX4, cation/H+ exchanger 4, CHX4, cation/H+ exchanger 4 |
-1.808 |
0.004 |
| 518 |
AT4G36540 |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-1.805 |
-0.606 |
| 519 |
AT1G54040 |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-1.804 |
0.123 |
| 520 |
AT4G15440 |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-1.799 |
0.267 |
| 521 |
AT5G11590 |
AT5G11590
|
TINY2, TINY2 |
-1.797 |
-0.284 |
| 522 |
AT5G61350 |
AT5G61350
|
[Protein kinase superfamily protein] |
-1.795 |
-0.284 |
| 523 |
AT1G58520 |
AT1G58520
|
RXW8 |
-1.794 |
no data |
| 524 |
AT5G46830 |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-1.793 |
-0.294 |
| 525 |
AT4G18970 |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.791 |
-0.418 |
| 526 |
AT2G34925 |
AT2G34925
|
CLE42, CLAVATA3/ESR-RELATED 42 |
-1.789 |
no data |
| 527 |
AT3G27025 |
AT3G27025
|
unknown |
-1.786 |
no data |
| 528 |
AT5G12050 |
AT5G12050
|
unknown |
-1.781 |
-0.252 |
| 529 |
AT3G15570 |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-1.778 |
-0.361 |
| 530 |
AT3G53420 |
AT3G53420
|
AtPIP2;1, PIP2, PLASMA MEMBRANE INTRINSIC PROTEIN 2, PIP2;1, PLASMA MEMBRANE INTRINSIC PROTEIN 2;1, PIP2A, plasma membrane intrinsic protein 2A |
-1.778 |
0.002 |
| 531 |
AT5G63180 |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-1.775 |
-0.338 |
| 532 |
AT2G37170 |
AT2G37170
|
PIP2;2, PLASMA MEMBRANE INTRINSIC PROTEIN 2;2, PIP2B, plasma membrane intrinsic protein 2 |
-1.771 |
no data |
| 533 |
AT5G62100 |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-1.770 |
-0.597 |
| 534 |
AT3G14810 |
AT3G14810
|
MSL5, mechanosensitive channel of small conductance-like 5 |
-1.768 |
no data |
| 535 |
AT4G38690 |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-1.763 |
-0.186 |
| 536 |
AT1G52342 |
AT1G52342
|
unknown |
-1.762 |
no data |
| 537 |
AT2G45830 |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-1.759 |
-0.025 |
| 538 |
AT1G78450 |
AT1G78450
|
[SOUL heme-binding family protein] |
-1.754 |
-0.432 |
| 539 |
AT1G50280 |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
-1.753 |
-0.195 |
| 540 |
AT4G27360 |
AT4G27360
|
[Dynein light chain type 1 family protein] |
-1.750 |
no data |
| 541 |
AT5G01520 |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-1.750 |
0.081 |
| 542 |
AT5G37300 |
AT5G37300
|
WSD1 |
-1.749 |
0.012 |
| 543 |
AT1G05690 |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
-1.748 |
-0.249 |
| 544 |
AT3G06770 |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-1.744 |
-0.566 |
| 545 |
AT3G11110 |
AT3G11110
|
[RING/U-box superfamily protein] |
-1.743 |
-0.210 |
| 546 |
AT4G01060 |
AT4G01060
|
CPL3, CAPRICE-like MYB3, ETC3, ENHANCER OF TRY AND CPC 3 |
-1.743 |
no data |
| 547 |
AT2G45560 |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
-1.743 |
-0.131 |
| 548 |
AT1G31173 |
AT1G31173
|
MIR167D, microRNA167D |
-1.743 |
no data |
| 549 |
AT5G49730 |
AT5G49730
|
ATFRO6, ferric reduction oxidase 6, FRO6, FRO6, ferric reduction oxidase 6 |
-1.731 |
no data |
| 550 |
AT1G51140 |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-1.730 |
-0.245 |
| 551 |
AT1G35290 |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-1.729 |
-0.032 |
| 552 |
AT1G55330 |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-1.725 |
-0.351 |
| 553 |
AT3G20820 |
AT3G20820
|
[Leucine-rich repeat (LRR) family protein] |
-1.722 |
-0.230 |
| 554 |
AT2G38544 |
AT2G38544
|
unknown |
-1.721 |
no data |
| 555 |
AT1G23080 |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-1.720 |
-0.330 |
| 556 |
AT3G61430 |
AT3G61430
|
ATPIP1, ARABIDOPSIS THALIANA PLASMA MEMBRANE INTRINSIC PROTEIN 1, PIP1, PLASMA MEMBRANE INTRINSIC PROTEIN 1, PIP1;1, PLASMA MEMBRANE INTRINSIC PROTEIN 1;1, PIP1A, plasma membrane intrinsic protein 1A |
-1.719 |
0.035 |
| 557 |
AT4G03060 |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-1.718 |
-0.190 |
| 558 |
AT4G22560 |
AT4G22560
|
unknown |
-1.717 |
-0.324 |
| 559 |
AT5G44680 |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-1.717 |
-0.604 |
| 560 |
AT4G18220 |
AT4G18220
|
[Drug/metabolite transporter superfamily protein] |
-1.713 |
no data |
| 561 |
AT1G04250 |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-1.713 |
-0.401 |
| 562 |
AT5G25460 |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-1.712 |
-0.330 |
| 563 |
AT5G01240 |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
-1.709 |
-0.354 |
| 564 |
AT5G23750 |
AT5G23750
|
[Remorin family protein] |
-1.707 |
-0.177 |
| 565 |
AT3G12610 |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-1.706 |
-0.829 |
| 566 |
AT1G19630 |
AT1G19630
|
CYP722A1, cytochrome P450, family 722, subfamily A, polypeptide 1 |
-1.705 |
-0.108 |
| 567 |
AT4G38860 |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-1.705 |
-0.523 |
| 568 |
AT2G21830 |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.701 |
-0.343 |
| 569 |
AT2G25625 |
AT2G25625
|
unknown |
-1.700 |
0.111 |
| 570 |
AT1G14280 |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-1.699 |
-0.328 |
| 571 |
AT3G26280 |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-1.694 |
0.180 |
| 572 |
AT5G15160 |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-1.691 |
-0.093 |
| 573 |
AT1G78070 |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-1.690 |
0.225 |
| 574 |
AT5G03760 |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-1.689 |
-0.254 |
| 575 |
AT2G18050 |
AT2G18050
|
HIS1-3, histone H1-3 |
-1.686 |
0.384 |
| 576 |
AT5G47610 |
AT5G47610
|
[RING/U-box superfamily protein] |
-1.686 |
-0.733 |
| 577 |
AT4G28780 |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.685 |
-0.621 |
| 578 |
AT1G13448 |
AT1G13448
|
other RNA |
-1.683 |
no data |
| 579 |
AT4G28790 |
AT4G28790
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.681 |
no data |
| 580 |
AT3G19850 |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-1.679 |
-0.438 |
| 581 |
AT1G70940 |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-1.679 |
-0.376 |
| 582 |
AT5G23020 |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-1.676 |
-0.311 |
| 583 |
AT3G45940 |
AT3G45940
|
[Glycosyl hydrolases family 31 protein] |
-1.675 |
-0.081 |
| 584 |
AT4G04750 |
AT4G04750
|
[Major facilitator superfamily protein] |
-1.673 |
-0.466 |
| 585 |
AT5G13930 |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-1.673 |
-0.402 |
| 586 |
AT1G32780 |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-1.671 |
-0.405 |
| 587 |
AT5G40380 |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-1.671 |
-0.166 |
| 588 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
-1.670 |
no data |
| 589 |
AT1G73600 |
AT1G73600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.666 |
no data |
| 590 |
AT2G01420 |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-1.663 |
-0.442 |
| 591 |
AT3G28070 |
AT3G28070
|
UMAMIT46, Usually multiple acids move in and out Transporters 46 |
-1.663 |
no data |
| 592 |
AT1G64640 |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-1.663 |
-0.491 |
| 593 |
AT1G18810 |
AT1G18810
|
[phytochrome kinase substrate-related] |
-1.663 |
-0.265 |
| 594 |
AT2G33380 |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-1.662 |
0.398 |
| 595 |
AT5G48900 |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-1.661 |
0.000 |
| 596 |
AT5G62140 |
AT5G62140
|
unknown |
-1.658 |
-0.407 |
| 597 |
AT1G22160 |
AT1G22160
|
unknown |
-1.656 |
-0.196 |
| 598 |
AT5G10570 |
AT5G10570
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.656 |
no data |
| 599 |
AT4G23020 |
AT4G23020
|
TRM11, TON1 Recruiting Motif 11 |
-1.655 |
no data |
| 600 |
AT1G28610 |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.655 |
-0.015 |