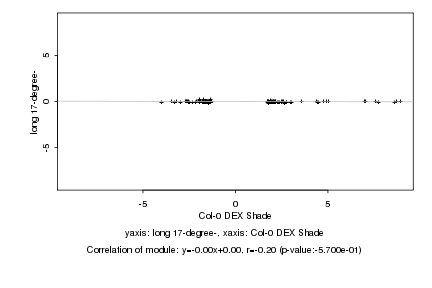
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 DEX Shade |
Log2 signal ratio
long 17-degree- |
| 1 |
AT4G27150 |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
8.902 |
0.017 |
| 2 |
AT4G27160 |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
8.699 |
0.018 |
| 3 |
AT4G27140 |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
8.564 |
-0.007 |
| 4 |
AT4G27170 |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
7.696 |
-0.017 |
| 5 |
AT1G03880 |
AT1G03880
|
CRB, CRUCIFERIN B, CRU2, cruciferin 2 |
7.544 |
0.013 |
| 6 |
AT4G28520 |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
7.016 |
0.032 |
| 7 |
AT1G47540 |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
6.964 |
0.028 |
| 8 |
AT4G25140 |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
5.017 |
0.034 |
| 9 |
AT4G31380 |
AT4G31380
|
FLP1, FPF1-like protein 1 |
4.955 |
no data |
| 10 |
AT5G54740 |
AT5G54740
|
SESA5, seed storage albumin 5 |
4.901 |
0.003 |
| 11 |
AT2G05580 |
AT2G05580
|
[Glycine-rich protein family] |
4.756 |
0.008 |
| 12 |
AT1G04180 |
AT1G04180
|
YUC9, YUCCA 9 |
4.447 |
-0.038 |
| 13 |
AT2G15010 |
AT2G15010
|
[Plant thionin] |
4.417 |
-0.017 |
| 14 |
AT4G26740 |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
4.337 |
0.034 |
| 15 |
AT2G44910 |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
3.537 |
0.003 |
| 16 |
AT3G21330 |
AT3G21330
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
3.429 |
no data |
| 17 |
AT4G32280 |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
3.028 |
-0.048 |
| 18 |
AT2G42870 |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
2.943 |
-0.048 |
| 19 |
ATCG00290 |
ATCG00290
|
TRNS.2 |
2.928 |
no data |
| 20 |
AT3G43850 |
AT3G43850
|
unknown |
2.754 |
no data |
| 21 |
AT4G16780 |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
2.735 |
-0.073 |
| 22 |
AT3G03830 |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
2.710 |
-0.058 |
| 23 |
AT3G62950 |
AT3G62950
|
[Thioredoxin superfamily protein] |
2.647 |
-0.179 |
| 24 |
AT5G65800 |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
2.596 |
0.013 |
| 25 |
AT4G35770 |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
2.588 |
-0.081 |
| 26 |
AT3G22961 |
AT3G22961
|
[Paired amphipathic helix (PAH2) superfamily protein] |
2.500 |
no data |
| 27 |
AT2G18700 |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
2.500 |
0.023 |
| 28 |
AT3G50800 |
AT3G50800
|
unknown |
2.490 |
no data |
| 29 |
AT4G27450 |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
2.469 |
-0.055 |
| 30 |
AT5G18050 |
AT5G18050
|
SAUR22, SMALL AUXIN UP RNA 22 |
2.460 |
no data |
| 31 |
AT5G44260 |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
2.448 |
0.011 |
| 32 |
AT5G18010 |
AT5G18010
|
SAUR19, small auxin up RNA 19 |
2.400 |
no data |
| 33 |
AT3G21320 |
AT3G21320
|
unknown |
2.364 |
no data |
| 34 |
AT5G12050 |
AT5G12050
|
unknown |
2.329 |
-0.059 |
| 35 |
AT4G37770 |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
2.321 |
-0.038 |
| 36 |
AT3G29370 |
AT3G29370
|
P1R3, P1R3 |
2.257 |
-0.036 |
| 37 |
AT2G46970 |
AT2G46970
|
PIL1, phytochrome interacting factor 3-like 1 |
2.154 |
no data |
| 38 |
AT1G26240 |
AT1G26240
|
[Proline-rich extensin-like family protein] |
2.121 |
-0.002 |
| 39 |
AT5G22920 |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
2.118 |
-0.089 |
| 40 |
AT3G47340 |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
2.117 |
0.001 |
| 41 |
AT1G02620 |
AT1G02620
|
[Ras-related small GTP-binding family protein] |
2.116 |
0.045 |
| 42 |
AT1G02660 |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
2.112 |
0.041 |
| 43 |
AT4G16000 |
AT4G16000
|
unknown |
2.107 |
-0.017 |
| 44 |
ATCG00350 |
ATCG00350
|
PSAA |
2.098 |
-0.008 |
| 45 |
AT1G02640 |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
2.091 |
-0.068 |
| 46 |
AT4G28720 |
AT4G28720
|
YUC8, YUCCA 8 |
2.090 |
0.011 |
| 47 |
AT4G34770 |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
2.083 |
-0.105 |
| 48 |
AT5G25640 |
AT5G25640
|
[Rhomboid-related intramembrane serine protease family protein] |
2.068 |
-0.023 |
| 49 |
AT1G02610 |
AT1G02610
|
[RING/FYVE/PHD zinc finger superfamily protein] |
2.056 |
no data |
| 50 |
AT5G43890 |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
2.042 |
0.043 |
| 51 |
AT3G62550 |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
2.038 |
-0.026 |
| 52 |
AT2G23270 |
AT2G23270
|
unknown |
2.012 |
0.033 |
| 53 |
AT3G48360 |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
2.007 |
-0.030 |
| 54 |
AT5G62280 |
AT5G62280
|
unknown |
2.003 |
-0.009 |
| 55 |
AT3G15450 |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
1.998 |
-0.073 |
| 56 |
AT5G47370 |
AT5G47370
|
HAT2 |
1.986 |
-0.053 |
| 57 |
AT5G39860 |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
1.983 |
-0.054 |
| 58 |
AT2G18010 |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
1.961 |
0.009 |
| 59 |
ATCG00720 |
ATCG00720
|
PETB, photosynthetic electron transfer B |
1.927 |
0.002 |
| 60 |
AT5G46240 |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
1.921 |
0.026 |
| 61 |
AT4G26288 |
AT4G26288
|
unknown |
1.896 |
no data |
| 62 |
AT1G49130 |
AT1G49130
|
BBX17, B-box domain protein 17 |
1.884 |
no data |
| 63 |
AT5G07010 |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.881 |
0.153 |
| 64 |
AT4G28085 |
AT4G28085
|
unknown |
1.870 |
-0.005 |
| 65 |
AT5G18060 |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
1.864 |
-0.088 |
| 66 |
ATCG00340 |
ATCG00340
|
PSAB |
1.862 |
-0.001 |
| 67 |
AT4G15990 |
AT4G15990
|
unknown |
1.853 |
no data |
| 68 |
AT1G53160 |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
1.850 |
0.041 |
| 69 |
AT3G17580 |
AT3G17580
|
unknown |
1.846 |
no data |
| 70 |
AT1G02340 |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
1.846 |
-0.018 |
| 71 |
AT4G36850 |
AT4G36850
|
[PQ-loop repeat family protein / transmembrane family protein] |
1.811 |
no data |
| 72 |
AT3G26200 |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
1.792 |
-0.103 |
| 73 |
AT5G56550 |
AT5G56550
|
ATOXS3, OXIDATIVE STRESS 3, OXS3, OXIDATIVE STRESS 3 |
1.787 |
no data |
| 74 |
AT3G23030 |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
1.778 |
-0.040 |
| 75 |
AT4G14130 |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
1.774 |
-0.038 |
| 76 |
AT1G11735 |
AT1G11735
|
MIR171B, microRNA171B |
1.763 |
no data |
| 77 |
AT3G29670 |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
1.762 |
0.026 |
| 78 |
AT5G66580 |
AT5G66580
|
unknown |
1.755 |
no data |
| 79 |
AT1G75250 |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
1.750 |
-0.112 |
| 80 |
AT1G11080 |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
1.750 |
-0.043 |
| 81 |
ATCG00040 |
ATCG00040
|
MATK, maturase K |
1.745 |
0.032 |
| 82 |
AT2G20670 |
AT2G20670
|
unknown |
1.743 |
-0.015 |
| 83 |
AT4G17460 |
AT4G17460
|
HAT1, JAB, JAIBA |
1.734 |
-0.105 |
| 84 |
AT5G51810 |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
1.731 |
0.024 |
| 85 |
AT4G36410 |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
1.728 |
-0.097 |
| 86 |
AT5G35777 |
AT5G35777
|
[copia-like retrotransposon family, has a 2.0e-67 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
1.727 |
no data |
| 87 |
AT1G69490 |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
1.722 |
0.055 |
| 88 |
AT4G15248 |
AT4G15248
|
BBX30, B-box domain protein 30 |
-5.733 |
no data |
| 89 |
AT4G34550 |
AT4G34550
|
unknown |
-4.555 |
no data |
| 90 |
AT2G15020 |
AT2G15020
|
unknown |
-4.010 |
-0.029 |
| 91 |
AT3G21150 |
AT3G21150
|
B-box 32 |
-3.503 |
no data |
| 92 |
AT5G20790 |
AT5G20790
|
unknown |
-3.476 |
0.038 |
| 93 |
AT2G36750 |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
-3.345 |
-0.013 |
| 94 |
AT2G27420 |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-3.232 |
0.017 |
| 95 |
AT3G21890 |
AT3G21890
|
BBX31, B-box domain protein 31 |
-3.015 |
-0.001 |
| 96 |
AT3G09450 |
AT3G09450
|
unknown |
-2.936 |
no data |
| 97 |
AT3G44450 |
AT3G44450
|
unknown |
-2.720 |
0.023 |
| 98 |
AT1G65870 |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-2.684 |
0.044 |
| 99 |
AT3G15310 |
AT3G15310
|
unknown |
-2.681 |
0.031 |
| 100 |
AT1G73010 |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-2.630 |
0.036 |
| 101 |
AT5G66740 |
AT5G66740
|
unknown |
-2.566 |
-0.009 |
| 102 |
AT3G17790 |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-2.542 |
0.053 |
| 103 |
AT4G27360 |
AT4G27360
|
[Dynein light chain type 1 family protein] |
-2.535 |
no data |
| 104 |
AT2G29090 |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-2.515 |
-0.031 |
| 105 |
AT2G21320 |
AT2G21320
|
BBX18, B-box domain protein 18 |
-2.361 |
-0.051 |
| 106 |
AT1G23205 |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-2.162 |
-0.030 |
| 107 |
AT4G29110 |
AT4G29110
|
unknown |
-2.159 |
no data |
| 108 |
AT5G46830 |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-2.117 |
0.003 |
| 109 |
AT1G66725 |
AT1G66725
|
MIR163, microRNA163 |
-2.036 |
no data |
| 110 |
AT1G48100 |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-1.981 |
0.129 |
| 111 |
AT1G01520 |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
-1.978 |
-0.006 |
| 112 |
AT3G52740 |
AT3G52740
|
unknown |
-1.963 |
0.052 |
| 113 |
AT1G61800 |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-1.955 |
0.232 |
| 114 |
AT1G51090 |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-1.916 |
0.047 |
| 115 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
-1.861 |
no data |
| 116 |
AT5G24155 |
AT5G24155
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-1.840 |
no data |
| 117 |
AT5G61350 |
AT5G61350
|
[Protein kinase superfamily protein] |
-1.812 |
-0.002 |
| 118 |
AT1G55910 |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-1.806 |
0.017 |
| 119 |
AT1G24580 |
AT1G24580
|
[RING/U-box superfamily protein] |
-1.764 |
no data |
| 120 |
AT5G42760 |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-1.763 |
0.023 |
| 121 |
AT3G22840 |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-1.755 |
0.234 |
| 122 |
AT3G43110 |
AT3G43110
|
unknown |
-1.751 |
-0.024 |
| 123 |
AT3G04140 |
AT3G04140
|
[Ankyrin repeat family protein] |
-1.745 |
no data |
| 124 |
AT4G01080 |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-1.740 |
0.051 |
| 125 |
AT5G20150 |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-1.735 |
0.075 |
| 126 |
AT1G60590 |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-1.731 |
0.010 |
| 127 |
AT2G11810 |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-1.706 |
-0.006 |
| 128 |
AT1G64500 |
AT1G64500
|
[Glutaredoxin family protein] |
-1.694 |
0.001 |
| 129 |
AT3G10150 |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-1.682 |
-0.026 |
| 130 |
AT1G66540 |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-1.657 |
-0.094 |
| 131 |
AT5G55340 |
AT5G55340
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-1.651 |
0.010 |
| 132 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-1.645 |
0.036 |
| 133 |
AT4G26850 |
AT4G26850
|
VTC2, vitamin c defective 2 |
-1.645 |
0.000 |
| 134 |
AT3G02040 |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-1.641 |
0.018 |
| 135 |
AT5G54470 |
AT5G54470
|
BBX29, B-box domain protein 29 |
-1.618 |
-0.005 |
| 136 |
AT1G17100 |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
-1.616 |
0.005 |
| 137 |
AT2G30766 |
AT2G30766
|
unknown |
-1.615 |
no data |
| 138 |
AT1G02820 |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-1.608 |
0.090 |
| 139 |
AT2G36800 |
AT2G36800
|
DOGT1, don-glucosyltransferase 1, UGT73C5, UDP-GLUCOSYL TRANSFERASE 73C5 |
-1.603 |
no data |
| 140 |
AT2G36630 |
AT2G36630
|
[Sulfite exporter TauE/SafE family protein] |
-1.602 |
-0.021 |
| 141 |
AT2G25680 |
AT2G25680
|
MOT1, molybdate transporter 1 |
-1.593 |
-0.009 |
| 142 |
AT4G14400 |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-1.579 |
-0.021 |
| 143 |
AT2G44940 |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-1.577 |
-0.042 |
| 144 |
AT1G63580 |
AT1G63580
|
[Receptor-like protein kinase-related family protein] |
-1.570 |
0.048 |
| 145 |
AT1G47400 |
AT1G47400
|
unknown |
-1.555 |
-0.082 |
| 146 |
AT1G20070 |
AT1G20070
|
unknown |
-1.531 |
0.038 |
| 147 |
AT3G14200 |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-1.525 |
-0.046 |
| 148 |
AT3G47430 |
AT3G47430
|
PEX11B, peroxin 11B |
-1.514 |
-0.010 |
| 149 |
AT5G01790 |
AT5G01790
|
unknown |
-1.513 |
0.001 |
| 150 |
AT5G17300 |
AT5G17300
|
RVE1, REVEILLE 1 |
-1.513 |
0.018 |
| 151 |
AT5G24150 |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-1.498 |
-0.138 |
| 152 |
AT1G55960 |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.484 |
-0.036 |
| 153 |
AT5G41900 |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-1.476 |
0.002 |
| 154 |
AT1G10640 |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-1.459 |
0.025 |
| 155 |
AT2G46790 |
AT2G46790
|
APRR9, Arabidopsis pseudo-response regulator 9, PRR9, pseudo-response regulator 9, TL1, TOC1-LIKE PROTEIN 1 |
-1.437 |
no data |
| 156 |
AT5G23730 |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
-1.436 |
0.010 |
| 157 |
AT3G47420 |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-1.425 |
0.053 |
| 158 |
AT2G29170 |
AT2G29170
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.413 |
no data |
| 159 |
AT5G01820 |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-1.394 |
-0.091 |
| 160 |
AT4G17090 |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-1.389 |
-0.013 |
| 161 |
AT1G06980 |
AT1G06980
|
unknown |
-1.387 |
0.001 |
| 162 |
AT4G16880 |
AT4G16880
|
[Leucine-rich repeat (LRR) family protein] |
-1.386 |
no data |
| 163 |
AT3G17609 |
AT3G17609
|
HYH, HY5-homolog |
-1.383 |
0.098 |
| 164 |
AT5G45850 |
AT5G45850
|
unknown |
-1.378 |
-0.010 |
| 165 |
AT1G16730 |
AT1G16730
|
unknown |
-1.377 |
-0.014 |
| 166 |
AT2G39710 |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-1.367 |
0.046 |
| 167 |
AT3G21670 |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-1.365 |
-0.037 |
| 168 |
AT1G56650 |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-1.365 |
0.269 |
| 169 |
AT5G54130 |
AT5G54130
|
[Calcium-binding endonuclease/exonuclease/phosphatase family] |
-1.361 |
0.082 |
| 170 |
AT1G64900 |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
-1.358 |
-0.011 |
| 171 |
AT1G20470 |
AT1G20470
|
SAUR60, SMALL AUXIN UPREGULATED RNA 60 |
-1.353 |
no data |
| 172 |
AT4G02410 |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-1.335 |
0.018 |
| 173 |
AT4G00165 |
AT4G00165
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.329 |
no data |