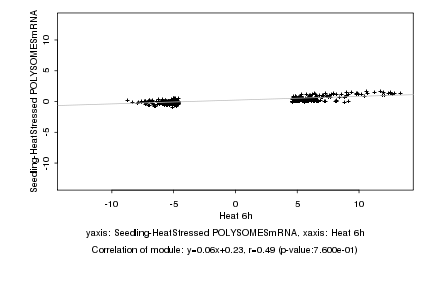
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Heat 6h |
Log2 signal ratio
Seedling-HeatStressed POLYSOMESmRNA |
| 1 |
AT1G53540 |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
13.325 |
1.411 |
| 2 |
AT4G10250 |
AT4G10250
|
ATHSP22.0 |
12.810 |
1.449 |
| 3 |
AT3G46230 |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
12.636 |
1.280 |
| 4 |
AT4G27670 |
AT4G27670
|
HSP21, heat shock protein 21 |
12.510 |
1.570 |
| 5 |
AT5G12020 |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
12.501 |
1.438 |
| 6 |
AT4G25200 |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
12.499 |
1.380 |
| 7 |
AT1G07400 |
AT1G07400
|
[HSP20-like chaperones superfamily protein] |
12.416 |
no data |
| 8 |
AT5G12030 |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
12.350 |
1.412 |
| 9 |
AT1G48710 |
AT1G48710
|
[copia-like retrotransposon family, has a 1.1e-252 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
12.232 |
no data |
| 10 |
AT5G59720 |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
11.985 |
1.029 |
| 11 |
AT1G71000 |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
11.963 |
1.594 |
| 12 |
AT2G29500 |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
11.880 |
1.004 |
| 13 |
AT1G52560 |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
11.690 |
1.711 |
| 14 |
AT3G09640 |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
11.242 |
1.606 |
| 15 |
AT4G12400 |
AT4G12400
|
Hop3, Hop3 |
10.618 |
1.418 |
| 16 |
AT2G26150 |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
10.592 |
1.640 |
| 17 |
AT3G12580 |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
10.406 |
0.910 |
| 18 |
AT1G03070 |
AT1G03070
|
AtLFG4, LFG4, LIFEGUARD 4 |
10.184 |
1.163 |
| 19 |
AT1G21940 |
AT1G21940
|
unknown |
10.098 |
no data |
| 20 |
AT5G48570 |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
9.945 |
1.192 |
| 21 |
AT5G51440 |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
9.806 |
1.404 |
| 22 |
AT3G32415 |
AT3G32415
|
[copia-like retrotransposon family, has a 8.1e-225 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
9.804 |
no data |
| 23 |
AT5G52640 |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
9.743 |
1.316 |
| 24 |
AT1G59860 |
AT1G59860
|
[HSP20-like chaperones superfamily protein] |
9.741 |
no data |
| 25 |
AT5G07330 |
AT5G07330
|
unknown |
9.728 |
1.281 |
| 26 |
AT3G59720 |
AT3G59720
|
[copia-like retrotransposon family, has a 1.1e-214 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
9.634 |
no data |
| 27 |
AT1G72660 |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
9.355 |
1.490 |
| 28 |
AT1G36630 |
AT1G36630
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 2.5e-213 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
9.188 |
no data |
| 29 |
AT3G61330 |
AT3G61330
|
[copia-like retrotransposon family, has a 1.1e-252 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
9.170 |
no data |
| 30 |
AT2G32120 |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
9.113 |
1.182 |
| 31 |
AT1G80660 |
AT1G80660
|
AHA9, H(+)-ATPase 9, HA9, H(+)-ATPase 9 |
9.102 |
0.088 |
| 32 |
AT2G21820 |
AT2G21820
|
unknown |
8.951 |
0.891 |
| 33 |
AT1G07500 |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
8.948 |
1.066 |
| 34 |
AT3G63350 |
AT3G63350
|
AT-HSFA7B, HSFA7B, HEAT SHOCK TRANSCRIPTION FACTOR A7B |
8.930 |
1.491 |
| 35 |
AT4G21320 |
AT4G21320
|
HSA32, HEAT-STRESS-ASSOCIATED 32 |
8.846 |
no data |
| 36 |
AT5G15250 |
AT5G15250
|
ATFTSH6, FTSH6, FTSH protease 6 |
8.784 |
0.711 |
| 37 |
AT3G59730 |
AT3G59730
|
LecRK-V.6, L-type lectin receptor kinase V.6 |
8.767 |
-0.024 |
| 38 |
AT1G58130 |
AT1G58130
|
[pseudogene] |
8.697 |
no data |
| 39 |
AT1G74310 |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
8.580 |
1.247 |
| 40 |
AT3G17110 |
AT3G17110
|
[pseudogene] |
8.381 |
0.775 |
| 41 |
AT1G02980 |
AT1G02980
|
ATCUL2, CUL2, cullin 2 |
8.173 |
0.300 |
| 42 |
AT3G07150 |
AT3G07150
|
unknown |
8.160 |
1.238 |
| 43 |
AT5G47600 |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
8.130 |
0.061 |
| 44 |
AT4G04170 |
AT4G04170
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.3e-87 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
8.057 |
0.033 |
| 45 |
AT1G16030 |
AT1G16030
|
Hsp70b, heat shock protein 70B |
7.893 |
1.281 |
| 46 |
AT5G64510 |
AT5G64510
|
TIN1, tunicamycin induced 1 |
7.857 |
1.396 |
| 47 |
AT4G19430 |
AT4G19430
|
unknown |
7.766 |
1.303 |
| 48 |
AT5G13205 |
AT5G13205
|
[copia-like retrotransposon family, has a 8.1e-252 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
7.755 |
no data |
| 49 |
AT2G11780 |
AT2G11780
|
[pseudogene] |
7.746 |
no data |
| 50 |
AT2G07731 |
AT2G07731
|
[pseudogene] |
7.741 |
no data |
| 51 |
AT3G01070 |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
7.664 |
0.771 |
| 52 |
AT1G17870 |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
7.583 |
1.007 |
| 53 |
AT3G53230 |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
7.556 |
0.826 |
| 54 |
AT2G37180 |
AT2G37180
|
PIP2;3, PLASMA MEMBRANE INTRINSIC PROTEIN 2;3, PIP2C, PLASMA MEMBRANE INTRINSIC PROTEIN 2C, RD28, RESPONSIVE TO DESICCATION 28 |
7.537 |
no data |
| 55 |
AT4G06566 |
AT4G06566
|
[gypsy-like retrotransposon family, has a 9.9e-101 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
7.522 |
no data |
| 56 |
AT5G19875 |
AT5G19875
|
unknown |
7.456 |
0.692 |
| 57 |
AT2G47180 |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
7.403 |
0.870 |
| 58 |
AT1G54050 |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
7.320 |
0.830 |
| 59 |
AT1G06030 |
AT1G06030
|
[pfkB-like carbohydrate kinase family protein] |
7.302 |
no data |
| 60 |
AT4G12530 |
AT4G12530
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
7.254 |
0.033 |
| 61 |
AT3G08970 |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
7.229 |
1.361 |
| 62 |
AT2G15810 |
AT2G15810
|
[Mutator-like transposase family, has a 1.5e-12 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
7.175 |
0.141 |
| 63 |
AT2G20560 |
AT2G20560
|
[DNAJ heat shock family protein] |
7.060 |
1.109 |
| 64 |
AT1G50390 |
AT1G50390
|
[pfkB-like carbohydrate kinase family protein] |
7.022 |
no data |
| 65 |
AT1G38430 |
AT1G38430
|
[gypsy-like retrotransposon family (Athila), has a 9.8e-109 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
7.017 |
no data |
| 66 |
AT5G27845 |
AT5G27845
|
[non-LTR retrotransposon family (LINE), has a 6.6e-32 P-value blast match to GB:AAA39398 ORF2 (Mus musculus) (LINE-element)] |
7.015 |
no data |
| 67 |
AT5G18340 |
AT5G18340
|
[ARM repeat superfamily protein] |
7.008 |
1.068 |
| 68 |
AT5G12110 |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
6.985 |
0.668 |
| 69 |
ATMG00630 |
ATMG00630
|
ORF110B |
6.939 |
0.732 |
| 70 |
ATMG00080 |
ATMG00080
|
RPL16, ribosomal protein L16 |
6.838 |
no data |
| 71 |
AT5G09930 |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
6.837 |
0.188 |
| 72 |
AT1G11265 |
AT1G11265
|
[copia-like retrotransposon family, has a 3.4e-253 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
6.802 |
no data |
| 73 |
AT4G23493 |
AT4G23493
|
unknown |
6.747 |
1.128 |
| 74 |
AT1G48700 |
AT1G48700
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
6.712 |
no data |
| 75 |
AT1G35730 |
AT1G35730
|
APUM9, pumilio 9, PUM9, pumilio 9 |
6.643 |
0.069 |
| 76 |
AT3G48240 |
AT3G48240
|
[Octicosapeptide/Phox/Bem1p family protein] |
6.639 |
no data |
| 77 |
AT5G57260 |
AT5G57260
|
CYP71B10, cytochrome P450, family 71, subfamily B, polypeptide 10 |
6.624 |
0.067 |
| 78 |
AT1G30070 |
AT1G30070
|
[SGS domain-containing protein] |
6.590 |
0.807 |
| 79 |
ATMG00070 |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
6.582 |
0.671 |
| 80 |
AT3G22090 |
AT3G22090
|
unknown |
6.551 |
0.951 |
| 81 |
AT2G01023 |
AT2G01023
|
unknown |
6.543 |
no data |
| 82 |
ATMG00060 |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
6.540 |
0.437 |
| 83 |
AT3G15340 |
AT3G15340
|
PPI2, proton pump interactor 2 |
6.517 |
no data |
| 84 |
AT3G03341 |
AT3G03341
|
unknown |
6.517 |
no data |
| 85 |
AT4G14080 |
AT4G14080
|
MEE48, maternal effect embryo arrest 48 |
6.503 |
0.056 |
| 86 |
AT3G41762 |
AT3G41762
|
unknown |
6.479 |
no data |
| 87 |
AT1G01940 |
AT1G01940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
6.464 |
0.434 |
| 88 |
AT2G07688 |
AT2G07688
|
[pseudogene] |
6.428 |
no data |
| 89 |
ATMG00090 |
ATMG00090
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
6.427 |
no data |
| 90 |
AT1G33500 |
AT1G33500
|
unknown |
6.419 |
0.315 |
| 91 |
AT3G24500 |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
6.406 |
0.778 |
| 92 |
AT2G23110 |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
6.398 |
1.283 |
| 93 |
AT1G44414 |
AT1G44414
|
unknown |
6.390 |
1.405 |
| 94 |
AT1G02065 |
AT1G02065
|
SPL8, squamosa promoter binding protein-like 8 |
6.388 |
no data |
| 95 |
AT1G35735 |
AT1G35735
|
[copia-like retrotransposon family, has a 7.7e-113 P-value blast match to GB:BAA22288 pol polyprotein (Ty1_Copia-element) (Oryza australiensis)GB:BAA22288 polyprotein (Ty1_Copia-element) (Oryza australiensis)gi|2443320|dbj|BAA22288.1| polyprotein (RIRE1) (Oryza australiensis) (Ty1_Copia-element)] |
6.385 |
no data |
| 96 |
ATMG00160 |
ATMG00160
|
COX2, cytochrome oxidase 2 |
6.362 |
0.647 |
| 97 |
AT4G36490 |
AT4G36490
|
ATSFH12, SEC14-LIKE 12, SFH12, SEC14-like 12 |
6.354 |
0.142 |
| 98 |
AT2G07783 |
AT2G07783
|
[pseudogene] |
6.347 |
no data |
| 99 |
AT5G22430 |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
6.319 |
0.022 |
| 100 |
AT2G07795 |
AT2G07795
|
unknown |
6.308 |
no data |
| 101 |
AT2G07733 |
AT2G07733
|
[pseudogene] |
6.289 |
no data |
| 102 |
AT2G46710 |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
6.288 |
0.027 |
| 103 |
AT1G21945 |
AT1G21945
|
[copia-like retrotransposon family, has a 1.7e-243 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
6.286 |
no data |
| 104 |
AT1G48730 |
AT1G48730
|
unknown |
6.237 |
0.051 |
| 105 |
ATMG01360 |
ATMG01360
|
COX1, cytochrome oxidase |
6.213 |
0.537 |
| 106 |
AT4G30460 |
AT4G30460
|
[glycine-rich protein] |
6.190 |
0.382 |
| 107 |
AT2G07682 |
AT2G07682
|
[gypsy-like retrotransposon family, has a 2.1e-65 P-value blast match to GB:AAD22153 polyprotein (Gypsy_Ty3-element) (Sorghum bicolor)] |
6.180 |
no data |
| 108 |
AT2G46240 |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
6.171 |
1.265 |
| 109 |
AT1G27710 |
AT1G27710
|
[Glycine-rich protein family] |
6.159 |
no data |
| 110 |
AT1G67360 |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
6.150 |
0.626 |
| 111 |
AT3G16050 |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
6.121 |
0.804 |
| 112 |
AT2G07677 |
AT2G07677
|
[auxin-responsive factor (ARF) -related, low similarity to auxin response factor 10 (Arabidopsis thaliana) GI:6165644] |
6.114 |
no data |
| 113 |
AT3G20190 |
AT3G20190
|
PRK4, pollen receptor like kinase 4 |
6.112 |
0.117 |
| 114 |
AT3G57880 |
AT3G57880
|
[Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase family protein] |
6.096 |
0.561 |
| 115 |
AT5G47830 |
AT5G47830
|
unknown |
6.095 |
0.717 |
| 116 |
AT2G19310 |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
6.087 |
0.556 |
| 117 |
AT2G11778 |
AT2G11778
|
unknown |
6.071 |
no data |
| 118 |
AT5G50790 |
AT5G50790
|
AtSWEET10, SWEET10 |
6.062 |
0.203 |
| 119 |
AT4G39361 |
AT4G39361
|
[snoRNA] |
6.049 |
no data |
| 120 |
ATMG00690 |
ATMG00690
|
ORF240A |
6.032 |
0.696 |
| 121 |
AT5G25450 |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
5.993 |
1.059 |
| 122 |
AT1G58140 |
AT1G58140
|
[copia-like retrotransposon family, has a 1.6e-253 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
5.985 |
no data |
| 123 |
AT5G05220 |
AT5G05220
|
unknown |
5.983 |
1.095 |
| 124 |
AT5G47610 |
AT5G47610
|
[RING/U-box superfamily protein] |
5.969 |
0.818 |
| 125 |
AT2G30000 |
AT2G30000
|
[PHF5-like protein] |
5.957 |
0.405 |
| 126 |
AT3G09350 |
AT3G09350
|
Fes1A, Fes1A |
5.925 |
0.903 |
| 127 |
AT4G29770 |
AT4G29770
|
unknown |
5.906 |
0.788 |
| 128 |
AT2G20870 |
AT2G20870
|
[cell wall protein precursor, putative] |
5.893 |
0.139 |
| 129 |
AT5G35320 |
AT5G35320
|
unknown |
5.879 |
0.749 |
| 130 |
AT3G24610 |
AT3G24610
|
[Galactose oxidase/kelch repeat superfamily protein] |
5.873 |
no data |
| 131 |
AT4G32212 |
AT4G32212
|
|
5.866 |
no data |
| 132 |
AT5G49920 |
AT5G49920
|
[Octicosapeptide/Phox/Bem1p family protein] |
5.852 |
no data |
| 133 |
AT3G19270 |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
5.847 |
0.566 |
| 134 |
AT4G06506 |
AT4G06506
|
[gypsy-like retrotransposon family (Athila), has a 3.1e-255 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
5.847 |
no data |
| 135 |
ATMG00400 |
ATMG00400
|
ORF157 |
5.835 |
0.258 |
| 136 |
AT5G43300 |
AT5G43300
|
AtGDPD3, GDPD3, glycerophosphodiester phosphodiesterase 3 |
5.835 |
no data |
| 137 |
AT5G52160 |
AT5G52160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
5.817 |
0.026 |
| 138 |
AT4G03320 |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
5.792 |
0.395 |
| 139 |
AT3G01600 |
AT3G01600
|
anac044, NAC domain containing protein 44, NAC044, NAC domain containing protein 44 |
5.785 |
no data |
| 140 |
AT2G46720 |
AT2G46720
|
HIC, HIGH CARBON DIOXIDE, KCS13, 3-ketoacyl-CoA synthase 13 |
5.782 |
no data |
| 141 |
AT4G04530 |
AT4G04530
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G27780.1)] |
5.776 |
0.020 |
| 142 |
AT2G07734 |
AT2G07734
|
[Alpha-L RNA-binding motif/Ribosomal protein S4 family protein] |
5.763 |
no data |
| 143 |
ATMG01220 |
ATMG01220
|
ORF113 |
5.754 |
no data |
| 144 |
AT2G07722 |
AT2G07722
|
unknown |
5.747 |
no data |
| 145 |
AT1G60970 |
AT1G60970
|
[SNARE-like superfamily protein] |
5.747 |
0.303 |
| 146 |
AT3G13227 |
AT3G13227
|
[serine-rich protein-related] |
5.738 |
no data |
| 147 |
AT2G07749 |
AT2G07749
|
[Mitovirus RNA-dependent RNA polymerase] |
5.737 |
no data |
| 148 |
AT2G01008 |
AT2G01008
|
unknown |
5.717 |
no data |
| 149 |
AT4G31877 |
AT4G31877
|
MIR156C, microRNA156C |
5.714 |
no data |
| 150 |
AT1G62510 |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
5.695 |
0.250 |
| 151 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
5.690 |
0.149 |
| 152 |
AT5G37670 |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
5.675 |
1.183 |
| 153 |
AT2G35743 |
AT2G35743
|
|
5.648 |
no data |
| 154 |
AT3G22840 |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
5.637 |
0.516 |
| 155 |
AT3G22100 |
AT3G22100
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
5.622 |
no data |
| 156 |
ATMG00560 |
ATMG00560
|
RPL2 |
5.613 |
no data |
| 157 |
AT3G13470 |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
5.605 |
0.423 |
| 158 |
ATMG01120 |
ATMG01120
|
NAD1, NADH DEHYDROGENASE 1, NAD1B, NADH dehydrogenase 1B |
5.597 |
no data |
| 159 |
AT4G19395 |
AT4G19395
|
MIR168, MICRORNA 168, MIR168A, microRNA168A |
5.590 |
no data |
| 160 |
AT2G07739 |
AT2G07739
|
[Ycf1 protein] |
5.587 |
no data |
| 161 |
AT4G18395 |
AT4G18395
|
unknown |
5.575 |
0.115 |
| 162 |
AT5G54165 |
AT5G54165
|
unknown |
5.551 |
no data |
| 163 |
AT4G39363 |
AT4G39363
|
[snoRNA] |
5.529 |
no data |
| 164 |
ATMG00040 |
ATMG00040
|
ORF315 |
5.523 |
no data |
| 165 |
AT3G30843 |
AT3G30843
|
unknown |
5.520 |
-0.045 |
| 166 |
AT3G10745 |
AT3G10745
|
MIR158A, microRNA158A |
5.515 |
no data |
| 167 |
AT1G48720 |
AT1G48720
|
unknown |
5.496 |
no data |
| 168 |
AT1G10220 |
AT1G10220
|
unknown |
5.483 |
no data |
| 169 |
ATMG01200 |
ATMG01200
|
ORF294 |
5.458 |
0.398 |
| 170 |
AT5G49700 |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
5.435 |
0.596 |
| 171 |
AT2G07835 |
AT2G07835
|
unknown |
5.429 |
no data |
| 172 |
AT2G07697 |
AT2G07697
|
[non-LTR retrotransposon family (LINE), has a 1.9e-23 P-value blast match to GB:NP_038605 L1 repeat, Tf subfamily, member 30 (LINE-element) (Mus musculus)] |
5.421 |
no data |
| 173 |
AT5G55835 |
AT5G55835
|
MIR156H, microRNA156H |
5.413 |
no data |
| 174 |
AT1G07330 |
AT1G07330
|
unknown |
5.409 |
0.207 |
| 175 |
AT5G29040 |
AT5G29040
|
[pseudogene] |
5.401 |
0.083 |
| 176 |
AT2G07719 |
AT2G07719
|
[Putative membrane lipoprotein] |
5.382 |
no data |
| 177 |
AT3G30844 |
AT3G30844
|
[copia-like retrotransposon family, has a 8.6e-95 P-value blast match to GB:CAA32025 ORF (Ty1_Copia-element) (Nicotiana tabacum)GB:CAA32025 ORF (Ty1_Copia-element) (Nicotiana tabacum)] |
5.376 |
no data |
| 178 |
AT2G22240 |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
5.366 |
0.514 |
| 179 |
AT2G07698 |
AT2G07698
|
[ATPase, F1 complex, alpha subunit protein] |
5.348 |
no data |
| 180 |
AT2G09187 |
AT2G09187
|
[gypsy-like retrotransposon family (Athila), has a 2.3e-226 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
5.341 |
no data |
| 181 |
AT2G25140 |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
5.341 |
0.950 |
| 182 |
AT5G58110 |
AT5G58110
|
[chaperone binding] |
5.337 |
0.268 |
| 183 |
AT2G07708 |
AT2G07708
|
unknown |
5.323 |
no data |
| 184 |
ATMG00650 |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
5.300 |
0.669 |
| 185 |
AT4G39364 |
AT4G39364
|
[snoRNA] |
5.291 |
no data |
| 186 |
AT5G01180 |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
5.285 |
1.148 |
| 187 |
AT1G11100 |
AT1G11100
|
[SNF2 domain-containing protein / helicase domain-containing protein / zinc finger protein-related] |
5.283 |
0.718 |
| 188 |
AT2G07717 |
AT2G07717
|
[pseudogene] |
5.281 |
no data |
| 189 |
AT1G56170 |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
5.264 |
0.592 |
| 190 |
AT2G07798 |
AT2G07798
|
unknown |
5.258 |
no data |
| 191 |
AT3G03270 |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
5.250 |
0.382 |
| 192 |
AT3G23990 |
AT3G23990
|
HSP60, heat shock protein 60, HSP60-3B, HEAT SHOCK PROTEIN 60-3B |
5.240 |
0.471 |
| 193 |
AT1G42050 |
AT1G42050
|
[pseudogene] |
5.218 |
no data |
| 194 |
AT2G07767 |
AT2G07767
|
[expressed protein, similar to putative non-LTR retroelement reverse transcriptase] |
5.218 |
no data |
| 195 |
AT1G62760 |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
5.206 |
0.902 |
| 196 |
ATMG01130 |
ATMG01130
|
ORF106F |
5.204 |
0.279 |
| 197 |
ATMG00810 |
ATMG00810
|
ORF240B |
5.197 |
no data |
| 198 |
ATMG01350 |
ATMG01350
|
ORF145C |
5.191 |
0.251 |
| 199 |
ATMG01080 |
ATMG01080
|
ATP9, mitochondrial F0-ATPase subunit 9 |
5.186 |
no data |
| 200 |
AT2G07683 |
AT2G07683
|
[copia-like retrotransposon family, has a 2.2e-110 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
5.177 |
no data |
| 201 |
AT1G07350 |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
5.174 |
0.739 |
| 202 |
AT5G48650 |
AT5G48650
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
5.171 |
0.085 |
| 203 |
AT1G13710 |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
5.167 |
0.223 |
| 204 |
AT5G56030 |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
5.142 |
0.436 |
| 205 |
AT5G62730 |
AT5G62730
|
[Major facilitator superfamily protein] |
5.134 |
0.236 |
| 206 |
AT5G09590 |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
5.131 |
0.805 |
| 207 |
AT3G44110 |
AT3G44110
|
ATJ, ATJ3, DNAJ homologue 3, J3 |
5.124 |
0.273 |
| 208 |
AT2G07686 |
AT2G07686
|
[gypsy-like retrotransposon family, has a 4.3e-28 P-value blast match to GB:CAB39733 rotease, reverse transcriptase, ribonuclease H, integrase (Gypsy_Ty3-element) (Drosophila buzzatii)] |
5.120 |
no data |
| 209 |
ATMG00130 |
ATMG00130
|
ORF121A |
5.110 |
0.168 |
| 210 |
ATMG00520 |
ATMG00520
|
MATR |
5.109 |
no data |
| 211 |
AT5G46610 |
AT5G46610
|
[Aluminium activated malate transporter family protein] |
5.107 |
no data |
| 212 |
AT3G29810 |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
5.103 |
0.730 |
| 213 |
AT1G21460 |
AT1G21460
|
AtSWEET1, SWEET1 |
5.091 |
0.316 |
| 214 |
AT3G12050 |
AT3G12050
|
[Aha1 domain-containing protein] |
5.082 |
0.509 |
| 215 |
AT4G33930 |
AT4G33930
|
[Cupredoxin superfamily protein] |
5.082 |
no data |
| 216 |
AT4G23680 |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
5.065 |
0.203 |
| 217 |
AT1G72100 |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
5.043 |
0.399 |
| 218 |
AT2G36750 |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
5.042 |
0.657 |
| 219 |
AT1G75750 |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
5.041 |
0.227 |
| 220 |
AT1G78400 |
AT1G78400
|
[Pectin lyase-like superfamily protein] |
5.029 |
0.107 |
| 221 |
AT2G07693 |
AT2G07693
|
[copia-like retrotransposon family, has a 6.2e-16 P-value blast match to GB:CAA37924 orf 2 (Ty1_Copia-element) (Arabidopsis thaliana)] |
5.026 |
0.062 |
| 222 |
AT2G07747 |
AT2G07747
|
[pseudogene] |
5.010 |
no data |
| 223 |
ATMG00640 |
ATMG00640
|
ORF25 |
5.009 |
0.610 |
| 224 |
AT3G51590 |
AT3G51590
|
LTP12, lipid transfer protein 12 |
5.008 |
0.077 |
| 225 |
AT2G07785 |
AT2G07785
|
[NADH dehydrogenase family protein] |
5.003 |
no data |
| 226 |
AT4G39366 |
AT4G39366
|
[snoRNA] |
4.991 |
no data |
| 227 |
AT4G28395 |
AT4G28395
|
A7, ANTHER 7, ATA7, ARABIDOPSIS THALIANA ANTHER 7 |
4.983 |
0.179 |
| 228 |
AT4G28390 |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
4.965 |
0.632 |
| 229 |
ATMG00140 |
ATMG00140
|
ORF167 |
4.955 |
0.095 |
| 230 |
AT3G10020 |
AT3G10020
|
unknown |
4.935 |
0.617 |
| 231 |
AT2G07599 |
AT2G07599
|
unknown |
4.930 |
no data |
| 232 |
AT1G33350 |
AT1G33350
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
4.927 |
0.036 |
| 233 |
AT2G35742 |
AT2G35742
|
[snoRNA] |
4.925 |
no data |
| 234 |
AT2G33590 |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
4.925 |
0.657 |
| 235 |
AT5G51105 |
AT5G51105
|
unknown |
4.922 |
no data |
| 236 |
AT5G41380 |
AT5G41380
|
[CCT motif family protein] |
4.920 |
no data |
| 237 |
AT3G25230 |
AT3G25230
|
ATFKBP62, FKBP62, FK506 BINDING PROTEIN 62, ROF1, rotamase FKBP 1 |
4.914 |
0.629 |
| 238 |
AT2G07725 |
AT2G07725
|
[Ribosomal L5P family protein] |
4.907 |
no data |
| 239 |
AT4G34850 |
AT4G34850
|
LAP5, LESS ADHESIVE POLLEN 5, PSKB, polyketide synthase B |
4.906 |
0.191 |
| 240 |
AT2G44170 |
AT2G44170
|
ATNMT2, ARABIDOPSIS N-MYRISTOYLTRANSFERASE 2, NMT2, N-myristoyltransferase 2 |
4.904 |
0.728 |
| 241 |
AT2G07706 |
AT2G07706
|
unknown |
4.892 |
no data |
| 242 |
AT5G13150 |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
4.890 |
0.397 |
| 243 |
AT5G58590 |
AT5G58590
|
RANBP1, RAN binding protein 1 |
4.878 |
0.478 |
| 244 |
AT4G32208 |
AT4G32208
|
[heat shock protein 70 (Hsp 70) family protein] |
4.874 |
no data |
| 245 |
ATMG00410 |
ATMG00410
|
ATP6, ATPASE SUBUNIT 6, ATP6-1, ATPase subunit 6-1 |
4.866 |
no data |
| 246 |
ATMG00670 |
ATMG00670
|
ORF275 |
4.857 |
0.306 |
| 247 |
ATMG00730 |
ATMG00730
|
COX3, cytochrome c oxidase subunit 3 |
4.853 |
no data |
| 248 |
AT5G66110 |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
4.818 |
0.636 |
| 249 |
AT3G13277 |
AT3G13277
|
other RNA |
4.817 |
no data |
| 250 |
AT5G05410 |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
4.813 |
0.909 |
| 251 |
ATMG00660 |
ATMG00660
|
ORF149 |
4.813 |
0.499 |
| 252 |
AT2G09840 |
AT2G09840
|
unknown |
4.810 |
0.109 |
| 253 |
AT4G11660 |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
4.806 |
0.937 |
| 254 |
AT2G07806 |
AT2G07806
|
unknown |
4.802 |
no data |
| 255 |
AT5G15845 |
AT5G15845
|
other RNA |
4.799 |
no data |
| 256 |
AT4G33860 |
AT4G33860
|
[Glycosyl hydrolase family 10 protein] |
4.798 |
no data |
| 257 |
AT1G07850 |
AT1G07850
|
unknown |
4.798 |
0.039 |
| 258 |
AT3G43862 |
AT3G43862
|
[Athila retroelement ORF2 -related, temporary automated functional assignment] |
4.795 |
no data |
| 259 |
AT2G07712 |
AT2G07712
|
[pseudogene] |
4.795 |
no data |
| 260 |
AT3G15605 |
AT3G15605
|
[nucleic acid binding] |
4.795 |
0.076 |
| 261 |
AT2G07775 |
AT2G07775
|
unknown |
4.790 |
no data |
| 262 |
AT2G07732 |
AT2G07732
|
[Ribulose bisphosphate carboxylase large chain, catalytic domain] |
4.785 |
no data |
| 263 |
AT2G01020 |
AT2G01020
|
[5.8SrRNA] |
4.783 |
no data |
| 264 |
ATMG00960 |
ATMG00960
|
CCB203 |
4.782 |
no data |
| 265 |
AT2G28056 |
AT2G28056
|
MIR172, MICRORNA 172, MIR172A, microRNA172A |
4.750 |
no data |
| 266 |
AT5G18065 |
AT5G18065
|
unknown |
4.748 |
no data |
| 267 |
AT4G16550 |
AT4G16550
|
[HSP20-like chaperone] |
4.746 |
no data |
| 268 |
ATMG01020 |
ATMG01020
|
ORF153B |
4.733 |
0.133 |
| 269 |
AT2G07787 |
AT2G07787
|
unknown |
4.731 |
no data |
| 270 |
AT1G69120 |
AT1G69120
|
AGL7, AGAMOUS-like 7, AP1, APETALA1, AtAP1 |
4.730 |
0.069 |
| 271 |
AT2G07671 |
AT2G07671
|
[ATP synthase subunit C family protein] |
4.727 |
no data |
| 272 |
AT1G79920 |
AT1G79920
|
AtHsp70-15, Hsp70-15, heat shock protein 70-15 |
4.726 |
no data |
| 273 |
AT4G24410 |
AT4G24410
|
unknown |
4.705 |
0.095 |
| 274 |
AT3G13784 |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
4.692 |
0.122 |
| 275 |
AT2G07687 |
AT2G07687
|
[Cytochrome c oxidase, subunit III] |
4.690 |
no data |
| 276 |
AT1G33160 |
AT1G33160
|
[pseudogene] |
4.685 |
no data |
| 277 |
AT1G30060 |
AT1G30060
|
[COP1-interacting protein-related] |
4.681 |
no data |
| 278 |
ATCG00100 |
ATCG00100
|
TRNG.1 |
4.680 |
no data |
| 279 |
AT1G66510 |
AT1G66510
|
[AAR2 protein family] |
4.678 |
0.875 |
| 280 |
AT1G58150 |
AT1G58150
|
unknown |
4.667 |
0.385 |
| 281 |
AT5G62080 |
AT5G62080
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
4.659 |
0.031 |
| 282 |
AT5G03720 |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
4.658 |
0.737 |
| 283 |
AT4G07942 |
AT4G07942
|
[pseudogene] |
4.658 |
no data |
| 284 |
ATMG01090 |
ATMG01090
|
ORF262 |
4.657 |
no data |
| 285 |
AT2G40580 |
AT2G40580
|
[Protein kinase superfamily protein] |
4.655 |
0.123 |
| 286 |
AT4G22485 |
AT4G22485
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
4.650 |
no data |
| 287 |
AT2G38325 |
AT2G38325
|
MIR390, MICRORNA 390, MIR390A, microRNA390A |
4.639 |
no data |
| 288 |
ATMG00150 |
ATMG00150
|
ORF116 |
4.632 |
0.130 |
| 289 |
ATMG00110 |
ATMG00110
|
ABCI2, ATP-binding cassette I2, CCB206, cytochrome C biogenesis 206 |
4.616 |
0.114 |
| 290 |
AT1G72260 |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
4.613 |
0.037 |
| 291 |
AT1G41835 |
AT1G41835
|
[copia-like retrotransposon family, has a 1.5e-96 P-value blast match to GB:BAA22288 pol polyprotein (Ty1_Copia-element) (Oryza australiensis)GB:BAA22288 polyprotein (Ty1_Copia-element) (Oryza australiensis)gi|2443320|dbj|BAA22288.1| polyprotein (RIRE1) (Oryza australiensis) (Ty1_Copia-element)] |
4.612 |
no data |
| 292 |
AT5G14020 |
AT5G14020
|
[Endosomal targeting BRO1-like domain-containing protein] |
4.611 |
0.553 |
| 293 |
AT5G32053 |
AT5G32053
|
[pseudogene] |
4.610 |
no data |
| 294 |
ATCG00070 |
ATCG00070
|
PSBK, photosystem II reaction center protein K precursor |
4.608 |
0.123 |
| 295 |
AT4G13580 |
AT4G13580
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
4.608 |
0.085 |
| 296 |
AT1G12740 |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
4.607 |
0.460 |
| 297 |
ATMG01390 |
ATMG01390
|
RRN18 |
4.607 |
no data |
| 298 |
AT5G13170 |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
4.605 |
-0.090 |
| 299 |
AT4G12960 |
AT4G12960
|
AtGILT, GILT, gamma-interferon-responsive lysosomal thiol reductase |
4.605 |
0.089 |
| 300 |
AT2G01520 |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
4.597 |
0.069 |
| 301 |
AT1G72920 |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-8.771 |
0.186 |
| 302 |
AT4G29610 |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-8.370 |
-0.154 |
| 303 |
AT1G72910 |
AT1G72910
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-8.180 |
no data |
| 304 |
AT2G40610 |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-7.973 |
-0.239 |
| 305 |
AT5G59870 |
AT5G59870
|
HTA6, histone H2A 6 |
-7.889 |
-0.151 |
| 306 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-7.681 |
0.064 |
| 307 |
AT1G24147 |
AT1G24147
|
unknown |
-7.663 |
no data |
| 308 |
AT2G15390 |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-7.610 |
-0.011 |
| 309 |
AT3G29000 |
AT3G29000
|
[Calcium-binding EF-hand family protein] |
-7.588 |
no data |
| 310 |
AT1G13470 |
AT1G13470
|
unknown |
-7.415 |
0.086 |
| 311 |
AT2G25735 |
AT2G25735
|
unknown |
-7.411 |
-0.048 |
| 312 |
AT5G61000 |
AT5G61000
|
ATRPA70D, RPA70D |
-7.310 |
-0.400 |
| 313 |
AT3G48650 |
AT3G48650
|
[pseudogene] |
-7.291 |
0.065 |
| 314 |
AT4G14400 |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-7.288 |
0.020 |
| 315 |
AT3G44260 |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
-7.264 |
-0.087 |
| 316 |
AT3G50770 |
AT3G50770
|
CML41, calmodulin-like 41 |
-7.119 |
-0.021 |
| 317 |
AT3G45930 |
AT3G45930
|
[Histone superfamily protein] |
-7.118 |
no data |
| 318 |
AT5G10390 |
AT5G10390
|
H3.1, histone 3.1 |
-7.098 |
-0.535 |
| 319 |
AT1G24140 |
AT1G24140
|
[Matrixin family protein] |
-7.060 |
0.112 |
| 320 |
AT2G14560 |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-7.045 |
0.120 |
| 321 |
AT3G20600 |
AT3G20600
|
NDR1, non race-specific disease resistance 1 |
-7.007 |
no data |
| 322 |
AT5G26920 |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-7.001 |
0.263 |
| 323 |
AT5G51190 |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
-6.979 |
0.188 |
| 324 |
AT3G05800 |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
-6.969 |
-0.612 |
| 325 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-6.930 |
0.098 |
| 326 |
AT3G59320 |
AT3G59320
|
unknown |
-6.924 |
no data |
| 327 |
AT1G07135 |
AT1G07135
|
[glycine-rich protein] |
-6.905 |
no data |
| 328 |
AT5G54490 |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-6.839 |
0.155 |
| 329 |
AT2G24600 |
AT2G24600
|
[Ankyrin repeat family protein] |
-6.736 |
0.205 |
| 330 |
AT2G37950 |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-6.725 |
-0.245 |
| 331 |
AT4G13570 |
AT4G13570
|
HTA4, histone H2A 4 |
-6.719 |
0.069 |
| 332 |
AT1G06760 |
AT1G06760
|
[winged-helix DNA-binding transcription factor family protein] |
-6.718 |
-0.201 |
| 333 |
AT1G20190 |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-6.708 |
-0.477 |
| 334 |
AT1G02610 |
AT1G02610
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-6.644 |
no data |
| 335 |
AT3G46320 |
AT3G46320
|
[Histone superfamily protein] |
-6.635 |
no data |
| 336 |
AT5G59350 |
AT5G59350
|
unknown |
-6.620 |
-0.766 |
| 337 |
AT5G39860 |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-6.571 |
-0.575 |
| 338 |
AT4G33467 |
AT4G33467
|
unknown |
-6.530 |
no data |
| 339 |
AT3G03190 |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-6.526 |
-0.665 |
| 340 |
AT2G26560 |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-6.464 |
0.100 |
| 341 |
AT1G54820 |
AT1G54820
|
[Protein kinase superfamily protein] |
-6.385 |
-0.475 |
| 342 |
AT2G44230 |
AT2G44230
|
unknown |
-6.333 |
-0.358 |
| 343 |
AT1G21130 |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-6.295 |
-0.003 |
| 344 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
-6.282 |
0.441 |
| 345 |
AT4G20780 |
AT4G20780
|
CML42, calmodulin like 42 |
-6.220 |
-0.349 |
| 346 |
AT5G67390 |
AT5G67390
|
unknown |
-6.184 |
-0.405 |
| 347 |
AT1G20693 |
AT1G20693
|
HMG BETA 1, HIGH MOBILITY GROUP BETA 1, HMGB2, high mobility group B2, NFD02, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 02, NFD2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP D 2 |
-6.155 |
-0.135 |
| 348 |
AT3G53730 |
AT3G53730
|
[Histone superfamily protein] |
-6.091 |
-0.205 |
| 349 |
AT1G10560 |
AT1G10560
|
ATPUB18, ARABIDOPSIS THALIANA PLANT U-BOX 18, PUB18, plant U-box 18 |
-6.053 |
0.122 |
| 350 |
AT5G02670 |
AT5G02670
|
unknown |
-6.034 |
-0.066 |
| 351 |
AT4G31840 |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-6.031 |
-0.308 |
| 352 |
AT1G26260 |
AT1G26260
|
CIB5, cryptochrome-interacting basic-helix-loop-helix 5 |
-6.005 |
-0.154 |
| 353 |
AT4G36500 |
AT4G36500
|
unknown |
-6.001 |
-0.127 |
| 354 |
AT2G30420 |
AT2G30420
|
ETC2, ENHANCER OF TRY AND CPC 2 |
-5.966 |
no data |
| 355 |
AT1G70420 |
AT1G70420
|
unknown |
-5.950 |
-0.336 |
| 356 |
AT5G60900 |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-5.944 |
0.059 |
| 357 |
AT1G65390 |
AT1G65390
|
ATPP2-A5, phloem protein 2 A5, PP2-A5, phloem protein 2 A5 |
-5.942 |
0.386 |
| 358 |
AT5G51390 |
AT5G51390
|
unknown |
-5.925 |
0.206 |
| 359 |
AT4G16870 |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-5.923 |
0.190 |
| 360 |
AT4G11890 |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-5.921 |
0.061 |
| 361 |
AT4G26255 |
AT4G26255
|
other RNA |
-5.907 |
no data |
| 362 |
AT1G22250 |
AT1G22250
|
unknown |
-5.895 |
no data |
| 363 |
AT3G27690 |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-5.861 |
0.016 |
| 364 |
AT2G30620 |
AT2G30620
|
[winged-helix DNA-binding transcription factor family protein] |
-5.854 |
-0.127 |
| 365 |
AT4G38850 |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-5.852 |
-0.429 |
| 366 |
AT5G56120 |
AT5G56120
|
unknown |
-5.839 |
no data |
| 367 |
AT5G66640 |
AT5G66640
|
DAR3, DA1-related protein 3 |
-5.833 |
0.091 |
| 368 |
AT1G72930 |
AT1G72930
|
AtTN10, TIR, toll/interleukin-1 receptor-like, TN10, TIR-nucleotide binding site family 10 |
-5.795 |
no data |
| 369 |
AT1G18810 |
AT1G18810
|
[phytochrome kinase substrate-related] |
-5.794 |
-0.530 |
| 370 |
AT3G09020 |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-5.787 |
0.047 |
| 371 |
AT3G62950 |
AT3G62950
|
[Thioredoxin superfamily protein] |
-5.785 |
0.093 |
| 372 |
AT3G10525 |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-5.782 |
-0.063 |
| 373 |
AT5G47500 |
AT5G47500
|
PME5, pectin methylesterase 5 |
-5.750 |
-0.022 |
| 374 |
AT3G13404 |
AT3G13404
|
unknown |
-5.750 |
no data |
| 375 |
AT5G15770 |
AT5G15770
|
AtGNA1, glucose-6-phosphate acetyltransferase 1, GNA1, glucose-6-phosphate acetyltransferase 1 |
-5.748 |
-0.560 |
| 376 |
AT4G16740 |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
-5.717 |
0.131 |
| 377 |
AT3G04640 |
AT3G04640
|
[glycine-rich protein] |
-5.714 |
0.260 |
| 378 |
AT5G43730 |
AT5G43730
|
RSG2, Resistance Silenced Gene 2 |
-5.703 |
0.023 |
| 379 |
AT5G65207 |
AT5G65207
|
unknown |
-5.695 |
no data |
| 380 |
AT3G55660 |
AT3G55660
|
ATROPGEF6, ROPGEF6, ROP (rho of plants) guanine nucleotide exchange factor 6 |
-5.690 |
-0.290 |
| 381 |
AT1G13650 |
AT1G13650
|
unknown |
-5.689 |
-0.077 |
| 382 |
AT4G17490 |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-5.673 |
0.195 |
| 383 |
AT2G25060 |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-5.672 |
-0.255 |
| 384 |
AT1G62560 |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-5.646 |
-0.435 |
| 385 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-5.635 |
no data |
| 386 |
AT4G03060 |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-5.623 |
-0.634 |
| 387 |
AT3G59330 |
AT3G59330
|
unknown |
-5.616 |
no data |
| 388 |
AT5G54145 |
AT5G54145
|
unknown |
-5.604 |
no data |
| 389 |
AT1G28370 |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-5.603 |
0.083 |
| 390 |
AT2G30424 |
AT2G30424
|
TCL2, TRICHOMELESS 2 |
-5.599 |
no data |
| 391 |
AT5G16570 |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-5.593 |
0.119 |
| 392 |
AT5G20030 |
AT5G20030
|
[Plant Tudor-like RNA-binding protein] |
-5.590 |
-0.073 |
| 393 |
AT4G11000 |
AT4G11000
|
[Ankyrin repeat family protein] |
-5.590 |
0.042 |
| 394 |
AT4G21400 |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-5.581 |
-0.032 |
| 395 |
AT2G36970 |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-5.570 |
-0.284 |
| 396 |
AT3G29035 |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-5.564 |
-0.188 |
| 397 |
AT2G30230 |
AT2G30230
|
unknown |
-5.543 |
no data |
| 398 |
AT3G16720 |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
-5.524 |
-0.010 |
| 399 |
AT3G53360 |
AT3G53360
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-5.513 |
0.010 |
| 400 |
AT3G48290 |
AT3G48290
|
CYP71A24, cytochrome P450, family 71, subfamily A, polypeptide 24 |
-5.512 |
no data |
| 401 |
AT2G40435 |
AT2G40435
|
unknown |
-5.499 |
-0.609 |
| 402 |
AT1G01120 |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-5.480 |
-0.088 |
| 403 |
AT3G05200 |
AT3G05200
|
ATL6, Arabidopsis toxicos en levadura 6 |
-5.470 |
0.122 |
| 404 |
AT3G45860 |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-5.462 |
0.090 |
| 405 |
AT3G03130 |
AT3G03130
|
unknown |
-5.460 |
-0.270 |
| 406 |
AT4G23030 |
AT4G23030
|
[MATE efflux family protein] |
-5.453 |
0.066 |
| 407 |
AT2G22200 |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-5.446 |
-0.303 |
| 408 |
AT4G33790 |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-5.440 |
0.014 |
| 409 |
AT1G61795 |
AT1G61795
|
[PAK-box/P21-Rho-binding family protein] |
-5.437 |
no data |
| 410 |
AT4G04410 |
AT4G04410
|
[copia-like retrotransposon family, has a 1.7e-176 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-5.428 |
0.084 |
| 411 |
AT5G13060 |
AT5G13060
|
ABAP1, ARMADILLO BTB protein 1 |
-5.426 |
no data |
| 412 |
AT1G35260 |
AT1G35260
|
MLP165, MLP-like protein 165 |
-5.424 |
-0.377 |
| 413 |
AT5G03552 |
AT5G03552
|
MIR822A, microRNA822A |
-5.416 |
no data |
| 414 |
AT2G34150 |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-5.410 |
-0.214 |
| 415 |
AT5G03995 |
AT5G03995
|
unknown |
-5.409 |
no data |
| 416 |
AT4G28310 |
AT4G28310
|
unknown |
-5.409 |
-0.470 |
| 417 |
AT5G18030 |
AT5G18030
|
[SAUR-like auxin-responsive protein family ] |
-5.403 |
no data |
| 418 |
AT3G61190 |
AT3G61190
|
BAP1, BON association protein 1 |
-5.401 |
0.089 |
| 419 |
AT2G04495 |
AT2G04495
|
unknown |
-5.396 |
no data |
| 420 |
AT1G29395 |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-5.395 |
0.080 |
| 421 |
AT5G48490 |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-5.393 |
-0.065 |
| 422 |
AT3G03840 |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-5.392 |
-0.120 |
| 423 |
AT1G62630 |
AT1G62630
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-5.386 |
no data |
| 424 |
AT1G23790 |
AT1G23790
|
unknown |
-5.372 |
-0.393 |
| 425 |
AT1G63230 |
AT1G63230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-5.370 |
no data |
| 426 |
AT5G02890 |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-5.370 |
-0.486 |
| 427 |
AT5G37940 |
AT5G37940
|
[Zinc-binding dehydrogenase family protein] |
-5.369 |
no data |
| 428 |
AT3G47820 |
AT3G47820
|
PUB39, PLANT U-BOX 39 |
-5.369 |
no data |
| 429 |
AT3G25600 |
AT3G25600
|
[Calcium-binding EF-hand family protein] |
-5.362 |
-0.076 |
| 430 |
AT1G65790 |
AT1G65790
|
ARK1, receptor kinase 1, RK1, receptor kinase 1 |
-5.348 |
no data |
| 431 |
AT2G35710 |
AT2G35710
|
PGSIP7, plant glycogenin-like starch initiation protein 7 |
-5.328 |
0.045 |
| 432 |
AT3G15680 |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-5.328 |
-0.351 |
| 433 |
AT2G44240 |
AT2G44240
|
unknown |
-5.328 |
0.061 |
| 434 |
AT1G30420 |
AT1G30420
|
ABCC11, ATP-binding cassette C11, ATMRP12, multidrug resistance-associated protein 12, MRP12, multidrug resistance-associated protein 12 |
-5.325 |
no data |
| 435 |
AT1G79410 |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-5.321 |
-0.475 |
| 436 |
AT4G30650 |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-5.316 |
-0.208 |
| 437 |
AT4G11330 |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-5.316 |
-0.200 |
| 438 |
AT5G39670 |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-5.312 |
0.066 |
| 439 |
AT3G23870 |
AT3G23870
|
unknown |
-5.310 |
0.075 |
| 440 |
AT5G10400 |
AT5G10400
|
H3.1, histone 3.1 |
-5.310 |
-0.286 |
| 441 |
AT5G43080 |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
-5.308 |
-0.175 |
| 442 |
AT3G48720 |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-5.305 |
-0.022 |
| 443 |
AT1G76955 |
AT1G76955
|
[Expressed protein] |
-5.301 |
no data |
| 444 |
AT2G39710 |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-5.296 |
-0.425 |
| 445 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-5.295 |
0.101 |
| 446 |
AT2G40750 |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-5.294 |
-0.061 |
| 447 |
AT4G39780 |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
-5.285 |
-0.309 |
| 448 |
AT5G58670 |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
-5.276 |
0.115 |
| 449 |
AT2G18328 |
AT2G18328
|
ATRL4, RAD-like 4, RL4, RAD-like 4 |
-5.276 |
no data |
| 450 |
AT3G05415 |
AT3G05415
|
[non-LTR retrotransposon family (LINE), has a 9.0e-48 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
-5.271 |
no data |
| 451 |
AT1G57590 |
AT1G57590
|
[Pectinacetylesterase family protein] |
-5.263 |
-0.214 |
| 452 |
AT3G45940 |
AT3G45940
|
[Glycosyl hydrolases family 31 protein] |
-5.256 |
0.070 |
| 453 |
AT1G58602 |
AT1G58602
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-5.254 |
no data |
| 454 |
AT3G52400 |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-5.239 |
0.243 |
| 455 |
AT1G23965 |
AT1G23965
|
unknown |
-5.238 |
no data |
| 456 |
AT2G25250 |
AT2G25250
|
unknown |
-5.233 |
-0.184 |
| 457 |
AT3G09550 |
AT3G09550
|
[Ankyrin repeat family protein] |
-5.227 |
0.002 |
| 458 |
AT1G79700 |
AT1G79700
|
WRI4, WRINKLED 4 |
-5.209 |
0.436 |
| 459 |
ATCG00180 |
ATCG00180
|
RPOC1 |
-5.201 |
0.232 |
| 460 |
AT3G19850 |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-5.194 |
-0.097 |
| 461 |
AT5G49015 |
AT5G49015
|
[Expressed protein] |
-5.191 |
no data |
| 462 |
AT5G05890 |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-5.191 |
-0.186 |
| 463 |
AT5G66230 |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-5.184 |
-0.333 |
| 464 |
AT2G46650 |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-5.182 |
-0.410 |
| 465 |
AT3G07195 |
AT3G07195
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-5.179 |
no data |
| 466 |
AT1G02820 |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-5.177 |
-0.336 |
| 467 |
AT5G21940 |
AT5G21940
|
unknown |
-5.173 |
no data |
| 468 |
AT1G55450 |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-5.170 |
0.079 |
| 469 |
AT2G44740 |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-5.157 |
-0.221 |
| 470 |
AT1G11210 |
AT1G11210
|
unknown |
-5.153 |
-0.404 |
| 471 |
AT3G19330 |
AT3G19330
|
unknown |
-5.150 |
-0.106 |
| 472 |
AT5G45700 |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-5.150 |
-0.881 |
| 473 |
AT5G25210 |
AT5G25210
|
unknown |
-5.149 |
-0.155 |
| 474 |
AT2G17880 |
AT2G17880
|
DJC24, DNA J protein C24 |
-5.139 |
0.272 |
| 475 |
AT2G12170 |
AT2G12170
|
unknown |
-5.131 |
0.085 |
| 476 |
AT3G22160 |
AT3G22160
|
JAV1, jasmonate-associated VQ motif gene 1 |
-5.125 |
-0.062 |
| 477 |
AT2G16750 |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-5.116 |
-0.077 |
| 478 |
AT3G27540 |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-5.114 |
-0.279 |
| 479 |
AT1G71970 |
AT1G71970
|
unknown |
-5.101 |
-0.087 |
| 480 |
AT1G69890 |
AT1G69890
|
unknown |
-5.094 |
-0.249 |
| 481 |
AT1G74200 |
AT1G74200
|
AtRLP16, receptor like protein 16, RLP16, receptor like protein 16 |
-5.088 |
no data |
| 482 |
AT2G28400 |
AT2G28400
|
unknown |
-5.087 |
0.378 |
| 483 |
AT3G28340 |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-5.083 |
0.168 |
| 484 |
AT2G17040 |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-5.079 |
0.373 |
| 485 |
AT4G29780 |
AT4G29780
|
unknown |
-5.077 |
0.648 |
| 486 |
AT3G50140 |
AT3G50140
|
unknown |
-5.074 |
0.016 |
| 487 |
AT1G21110 |
AT1G21110
|
IGMT3, indole glucosinolate O-methyltransferase 3 |
-5.058 |
no data |
| 488 |
AT5G50740 |
AT5G50740
|
[Heavy metal transport/detoxification superfamily protein ] |
-5.055 |
-0.243 |
| 489 |
AT2G27660 |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-5.055 |
-0.160 |
| 490 |
AT3G13403 |
AT3G13403
|
[Defensin-like (DEFL) family protein] |
-5.045 |
no data |
| 491 |
AT2G17450 |
AT2G17450
|
RHA3A, RING-H2 finger A3A |
-5.038 |
-0.064 |
| 492 |
AT1G72900 |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-5.030 |
0.496 |
| 493 |
AT5G22250 |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-5.030 |
-0.157 |
| 494 |
AT1G21120 |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
-5.023 |
0.081 |
| 495 |
AT1G71030 |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-5.023 |
-0.063 |
| 496 |
AT4G36670 |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-5.022 |
-0.100 |
| 497 |
AT1G56130 |
AT1G56130
|
[Leucine-rich repeat transmembrane protein kinase] |
-5.020 |
no data |
| 498 |
AT1G62520 |
AT1G62520
|
unknown |
-5.018 |
0.185 |
| 499 |
AT3G05936 |
AT3G05936
|
unknown |
-4.999 |
no data |
| 500 |
AT1G16060 |
AT1G16060
|
ADAP, ARIA-interacting double AP2 domain protein, WRI3, WRINKLED 3 |
-4.995 |
-0.371 |
| 501 |
AT1G51700 |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-4.993 |
-0.134 |
| 502 |
AT1G68840 |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-4.989 |
0.101 |
| 503 |
AT4G15233 |
AT4G15233
|
ABCG42, ATP-binding cassette G42 |
-4.989 |
no data |
| 504 |
AT4G13560 |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-4.982 |
0.139 |
| 505 |
AT1G13410 |
AT1G13410
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-4.979 |
-0.131 |
| 506 |
AT5G27420 |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-4.977 |
0.558 |
| 507 |
AT4G01740 |
AT4G01740
|
[Cysteine/Histidine-rich C1 domain family protein] |
-4.973 |
-0.089 |
| 508 |
AT1G69530 |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-4.969 |
-0.491 |
| 509 |
AT4G27280 |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-4.966 |
0.309 |
| 510 |
AT1G36940 |
AT1G36940
|
unknown |
-4.948 |
0.126 |
| 511 |
AT3G28510 |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-4.943 |
0.000 |
| 512 |
AT2G40475 |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-4.936 |
-0.473 |
| 513 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
-4.936 |
no data |
| 514 |
AT3G12870 |
AT3G12870
|
unknown |
-4.934 |
-0.381 |
| 515 |
AT2G11810 |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-4.922 |
0.061 |
| 516 |
AT1G49500 |
AT1G49500
|
unknown |
-4.922 |
-0.039 |
| 517 |
AT4G30290 |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-4.920 |
0.093 |
| 518 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-4.915 |
0.534 |
| 519 |
AT1G05675 |
AT1G05675
|
[UDP-Glycosyltransferase superfamily protein] |
-4.914 |
no data |
| 520 |
AT3G52710 |
AT3G52710
|
unknown |
-4.914 |
-0.258 |
| 521 |
AT5G44005 |
AT5G44005
|
unknown |
-4.907 |
no data |
| 522 |
AT4G40065 |
AT4G40065
|
other RNA |
-4.907 |
no data |
| 523 |
AT4G38860 |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-4.902 |
-0.402 |
| 524 |
AT1G17147 |
AT1G17147
|
[VQ motif-containing protein] |
-4.901 |
no data |
| 525 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
-4.897 |
0.180 |
| 526 |
AT1G25560 |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-4.896 |
0.029 |
| 527 |
AT2G20142 |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-4.896 |
0.155 |
| 528 |
AT5G25190 |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-4.896 |
-0.057 |
| 529 |
AT3G52740 |
AT3G52740
|
unknown |
-4.894 |
-0.490 |
| 530 |
AT5G39680 |
AT5G39680
|
EMB2744, EMBRYO DEFECTIVE 2744 |
-4.894 |
-0.022 |
| 531 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-4.893 |
0.283 |
| 532 |
AT1G17430 |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-4.893 |
-0.052 |
| 533 |
AT1G61120 |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-4.892 |
0.023 |
| 534 |
AT2G15080 |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-4.889 |
-0.090 |
| 535 |
AT5G49525 |
AT5G49525
|
unknown |
-4.887 |
no data |
| 536 |
AT2G03590 |
AT2G03590
|
ATUPS1, ureide permease 1, UPS1, ureide permease 1 |
-4.887 |
no data |
| 537 |
AT1G58225 |
AT1G58225
|
unknown |
-4.882 |
no data |
| 538 |
AT5G10570 |
AT5G10570
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-4.882 |
no data |
| 539 |
AT1G49780 |
AT1G49780
|
PUB26, plant U-box 26 |
-4.877 |
0.089 |
| 540 |
AT5G20110 |
AT5G20110
|
[Dynein light chain type 1 family protein] |
-4.868 |
-0.172 |
| 541 |
AT1G29720 |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-4.855 |
-0.189 |
| 542 |
AT5G47230 |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-4.854 |
-0.115 |
| 543 |
AT3G01290 |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-4.853 |
0.240 |
| 544 |
AT1G74930 |
AT1G74930
|
ORA47 |
-4.850 |
0.015 |
| 545 |
ATCG00190 |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
-4.850 |
0.044 |
| 546 |
AT2G18210 |
AT2G18210
|
unknown |
-4.844 |
-0.346 |
| 547 |
AT3G44420 |
AT3G44420
|
[pseudogene] |
-4.844 |
no data |
| 548 |
AT1G35230 |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-4.844 |
0.076 |
| 549 |
AT3G59765 |
AT3G59765
|
other RNA |
-4.836 |
no data |
| 550 |
AT4G11370 |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
-4.833 |
-0.156 |
| 551 |
AT2G44490 |
AT2G44490
|
BGLU26, BETA GLUCOSIDASE 26, PEN2, PENETRATION 2 |
-4.828 |
-0.096 |
| 552 |
AT4G18422 |
AT4G18422
|
unknown |
-4.827 |
no data |
| 553 |
AT1G51270 |
AT1G51270
|
[structural molecules] |
-4.827 |
0.152 |
| 554 |
AT5G64660 |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
-4.818 |
-0.162 |
| 555 |
AT5G48657 |
AT5G48657
|
[defense protein-related] |
-4.817 |
no data |
| 556 |
AT5G65360 |
AT5G65360
|
H3.1, histone 3.1 |
-4.814 |
-0.203 |
| 557 |
AT2G03540 |
AT2G03540
|
unknown |
-4.813 |
0.145 |
| 558 |
AT3G58990 |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-4.804 |
-0.596 |
| 559 |
AT2G28720 |
AT2G28720
|
[Histone superfamily protein] |
-4.800 |
-0.655 |
| 560 |
AT1G62670 |
AT1G62670
|
RPF2, rna processing factor 2 |
-4.799 |
-0.035 |
| 561 |
AT5G50335 |
AT5G50335
|
unknown |
-4.799 |
-0.248 |
| 562 |
AT3G54150 |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-4.798 |
-0.021 |
| 563 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
-4.793 |
0.063 |
| 564 |
AT5G48310 |
AT5G48310
|
unknown |
-4.787 |
-0.094 |
| 565 |
AT4G23130 |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-4.785 |
0.028 |
| 566 |
AT4G13572 |
AT4G13572
|
unknown |
-4.783 |
no data |
| 567 |
AT2G42110 |
AT2G42110
|
unknown |
-4.782 |
-0.406 |
| 568 |
AT5G11070 |
AT5G11070
|
unknown |
-4.780 |
0.193 |
| 569 |
AT4G04450 |
AT4G04450
|
AtWRKY42, WRKY42 |
-4.779 |
-0.002 |
| 570 |
AT3G02640 |
AT3G02640
|
unknown |
-4.773 |
-0.345 |
| 571 |
AT4G34790 |
AT4G34790
|
SAUR3, SMALL AUXIN UPREGULATED RNA 3 |
-4.771 |
no data |
| 572 |
AT5G06150 |
AT5G06150
|
CYC1BAT, CYCB1;2, cyclin B 1;2 |
-4.770 |
no data |
| 573 |
AT1G52450 |
AT1G52450
|
[Ubiquitin carboxyl-terminal hydrolase-related protein] |
-4.769 |
no data |
| 574 |
AT4G37610 |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-4.766 |
0.185 |
| 575 |
AT5G65610 |
AT5G65610
|
unknown |
-4.763 |
-0.080 |
| 576 |
AT2G39705 |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-4.761 |
-0.268 |
| 577 |
AT5G55520 |
AT5G55520
|
unknown |
-4.754 |
-0.239 |
| 578 |
AT5G44820 |
AT5G44820
|
[Nucleotide-diphospho-sugar transferase family protein] |
-4.747 |
0.068 |
| 579 |
AT3G25100 |
AT3G25100
|
CDC45, cell division cycle 45 |
-4.735 |
-0.146 |
| 580 |
AT5G62630 |
AT5G62630
|
HIPL2, hipl2 protein precursor |
-4.731 |
-0.144 |
| 581 |
AT1G44110 |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
-4.726 |
-0.475 |
| 582 |
AT1G26762 |
AT1G26762
|
unknown |
-4.725 |
no data |
| 583 |
AT2G38790 |
AT2G38790
|
unknown |
-4.725 |
0.013 |
| 584 |
AT1G73805 |
AT1G73805
|
SARD1, SAR Deficient 1 |
-4.725 |
0.156 |
| 585 |
AT4G15215 |
AT4G15215
|
ABCG41, ATP-binding cassette G41, ATPDR13, PLEIOTROPIC DRUG RESISTANCE 13, PDR13, pleiotropic drug resistance 13 |
-4.723 |
no data |
| 586 |
AT3G12170 |
AT3G12170
|
[Chaperone DnaJ-domain superfamily protein] |
-4.703 |
-0.113 |
| 587 |
AT4G13395 |
AT4G13395
|
DVL10, DEVIL 10, RTFL12, ROTUNDIFOLIA like 12 |
-4.698 |
no data |
| 588 |
AT5G41590 |
AT5G41590
|
unknown |
-4.694 |
no data |
| 589 |
AT5G60680 |
AT5G60680
|
unknown |
-4.688 |
0.154 |
| 590 |
AT1G19200 |
AT1G19200
|
unknown |
-4.685 |
-0.019 |
| 591 |
AT1G76410 |
AT1G76410
|
ATL8 |
-4.684 |
-0.045 |
| 592 |
AT3G28945 |
AT3G28945
|
[non-LTR retrotransposon family (LINE), has a 2.5e-29 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-4.682 |
no data |
| 593 |
AT2G32030 |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-4.680 |
0.528 |
| 594 |
AT3G15210 |
AT3G15210
|
ATERF-4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ATERF4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ERF4, ethylene responsive element binding factor 4, RAP2.5, RELATED TO AP2 5 |
-4.676 |
0.159 |
| 595 |
AT1G12775 |
AT1G12775
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-4.674 |
0.046 |
| 596 |
AT3G23480 |
AT3G23480
|
[Cyclopropane-fatty-acyl-phospholipid synthase] |
-4.673 |
no data |
| 597 |
AT5G03230 |
AT5G03230
|
unknown |
-4.663 |
-0.215 |
| 598 |
AT1G04020 |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-4.663 |
-0.211 |
| 599 |
AT3G63210 |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-4.661 |
-0.208 |
| 600 |
AT3G09540 |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-4.657 |
-0.398 |