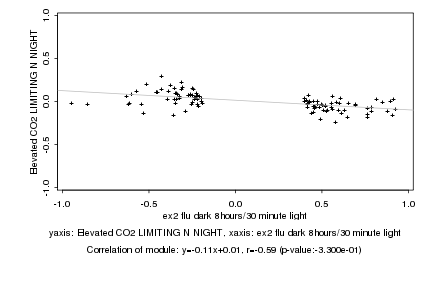
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ex2 flu dark 8hours/30 minute light |
Log2 signal ratio
Elevated CO2 LIMITING N NIGHT |
| 1 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.920 |
-0.089 |
| 2 |
256442_at |
AT3G10930
|
unknown |
0.909 |
0.028 |
| 3 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.906 |
-0.151 |
| 4 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.892 |
0.006 |
| 5 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.874 |
-0.115 |
| 6 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.847 |
-0.009 |
| 7 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.814 |
0.033 |
| 8 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.781 |
-0.114 |
| 9 |
263182_at |
AT1G05575
|
unknown |
0.781 |
-0.060 |
| 10 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
0.763 |
-0.078 |
| 11 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.760 |
-0.176 |
| 12 |
247047_at |
AT5G66650
|
unknown |
0.759 |
-0.144 |
| 13 |
246584_at |
AT5G14730
|
unknown |
0.693 |
-0.028 |
| 14 |
245755_at |
AT1G35210
|
unknown |
0.691 |
-0.043 |
| 15 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.649 |
-0.012 |
| 16 |
253044_at |
AT4G37290
|
unknown |
0.643 |
-0.175 |
| 17 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.629 |
-0.103 |
| 18 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.610 |
-0.139 |
| 19 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.602 |
0.040 |
| 20 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.600 |
-0.022 |
| 21 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.592 |
-0.094 |
| 22 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.581 |
-0.001 |
| 23 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.578 |
-0.233 |
| 24 |
254408_at |
AT4G21390
|
B120 |
0.557 |
-0.083 |
| 25 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.556 |
0.069 |
| 26 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.553 |
-0.020 |
| 27 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.551 |
-0.067 |
| 28 |
253643_at |
AT4G29780
|
unknown |
0.527 |
-0.096 |
| 29 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.526 |
-0.111 |
| 30 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.519 |
-0.046 |
| 31 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.519 |
-0.047 |
| 32 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.506 |
-0.102 |
| 33 |
250098_at |
AT5G17350
|
unknown |
0.500 |
-0.036 |
| 34 |
259979_at |
AT1G76600
|
unknown |
0.496 |
-0.029 |
| 35 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.486 |
-0.202 |
| 36 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.481 |
-0.061 |
| 37 |
245041_at |
AT2G26530
|
AR781 |
0.474 |
-0.026 |
| 38 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.473 |
0.009 |
| 39 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.462 |
-0.067 |
| 40 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.454 |
-0.051 |
| 41 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.453 |
-0.070 |
| 42 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.448 |
-0.120 |
| 43 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.447 |
0.003 |
| 44 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.447 |
-0.053 |
| 45 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.437 |
-0.131 |
| 46 |
267623_at |
AT2G39650
|
unknown |
0.426 |
-0.011 |
| 47 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.426 |
-0.001 |
| 48 |
247177_at |
AT5G65300
|
unknown |
0.423 |
0.010 |
| 49 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.417 |
0.075 |
| 50 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.416 |
-0.033 |
| 51 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.416 |
-0.058 |
| 52 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
0.411 |
-0.019 |
| 53 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.406 |
0.015 |
| 54 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
0.397 |
0.001 |
| 55 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.396 |
0.041 |
| 56 |
258027_at |
AT3G19515
|
unknown |
-0.952 |
-0.015 |
| 57 |
256617_at |
AT3G22240
|
unknown |
-0.860 |
-0.025 |
| 58 |
258424_at |
AT3G16750
|
unknown |
-0.630 |
0.062 |
| 59 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.624 |
-0.023 |
| 60 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.614 |
-0.018 |
| 61 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.604 |
0.090 |
| 62 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-0.573 |
0.117 |
| 63 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.547 |
-0.025 |
| 64 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.532 |
-0.132 |
| 65 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-0.517 |
0.200 |
| 66 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.460 |
0.106 |
| 67 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.460 |
0.105 |
| 68 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.456 |
0.116 |
| 69 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.433 |
0.292 |
| 70 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.431 |
0.142 |
| 71 |
257067_at |
AT3G18270
|
pseudogene |
-0.398 |
0.027 |
| 72 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.391 |
0.121 |
| 73 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.378 |
0.190 |
| 74 |
249454_at |
AT5G39520
|
unknown |
-0.363 |
-0.155 |
| 75 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.357 |
0.029 |
| 76 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.356 |
0.154 |
| 77 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.349 |
-0.016 |
| 78 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.348 |
0.096 |
| 79 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.343 |
0.034 |
| 80 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.343 |
0.095 |
| 81 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
-0.336 |
0.093 |
| 82 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.328 |
0.038 |
| 83 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.325 |
0.067 |
| 84 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.315 |
0.143 |
| 85 |
256883_at |
AT3G26440
|
unknown |
-0.312 |
0.221 |
| 86 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.308 |
0.173 |
| 87 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.293 |
-0.107 |
| 88 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.277 |
0.073 |
| 89 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.272 |
0.074 |
| 90 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.262 |
0.080 |
| 91 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
-0.262 |
0.087 |
| 92 |
258050_at |
AT3G16200
|
unknown |
-0.255 |
-0.030 |
| 93 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.253 |
0.153 |
| 94 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.251 |
-0.001 |
| 95 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
-0.251 |
0.071 |
| 96 |
245401_at |
AT4G17670
|
unknown |
-0.248 |
0.149 |
| 97 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.242 |
0.053 |
| 98 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.239 |
0.027 |
| 99 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.228 |
0.069 |
| 100 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-0.228 |
0.039 |
| 101 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.227 |
0.102 |
| 102 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.225 |
-0.026 |
| 103 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.224 |
0.034 |
| 104 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
-0.222 |
0.073 |
| 105 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.219 |
-0.053 |
| 106 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.212 |
0.059 |
| 107 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.202 |
0.008 |
| 108 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
-0.200 |
0.041 |
| 109 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.195 |
-0.017 |