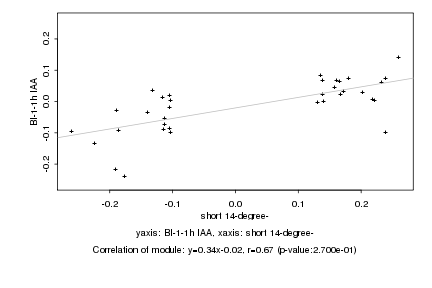
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
short 14-degree- |
Log2 signal ratio
BI-1-1h IAA |
| 1 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.259 |
0.141 |
| 2 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.238 |
-0.097 |
| 3 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.238 |
0.075 |
| 4 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.231 |
0.061 |
| 5 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.220 |
0.004 |
| 6 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.218 |
0.007 |
| 7 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.202 |
0.030 |
| 8 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.179 |
0.075 |
| 9 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.171 |
0.034 |
| 10 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.166 |
0.024 |
| 11 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.164 |
0.064 |
| 12 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.160 |
0.070 |
| 13 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.157 |
0.047 |
| 14 |
258166_at |
AT3G21540
|
[transducin family protein / WD-40 repeat family protein] |
0.139 |
0.003 |
| 15 |
255672_at |
AT4G00310
|
EDA8, EMBRYO SAC DEVELOPMENT ARREST 8, MEE46, MATERNAL EFFECT EMBRYO ARREST 46 |
0.138 |
0.023 |
| 16 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
0.137 |
0.068 |
| 17 |
251922_at |
AT3G54030
|
BSK6, brassinosteroid-signaling kinase 6 |
0.135 |
0.086 |
| 18 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.130 |
-0.001 |
| 19 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.262 |
-0.093 |
| 20 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.225 |
-0.134 |
| 21 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.192 |
-0.216 |
| 22 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.190 |
-0.026 |
| 23 |
246487_at |
AT5G16030
|
unknown |
-0.187 |
-0.091 |
| 24 |
256455_at |
AT1G75190
|
unknown |
-0.177 |
-0.239 |
| 25 |
254851_at |
AT4G12010
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.141 |
-0.032 |
| 26 |
264369_at |
AT1G70430
|
[Protein kinase superfamily protein] |
-0.132 |
0.038 |
| 27 |
258935_at |
AT3G10120
|
unknown |
-0.117 |
0.014 |
| 28 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.116 |
-0.089 |
| 29 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
-0.114 |
-0.071 |
| 30 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.113 |
-0.053 |
| 31 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.105 |
0.022 |
| 32 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.105 |
-0.085 |
| 33 |
259181_at |
AT3G01690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.105 |
-0.018 |
| 34 |
255854_at |
AT1G67050
|
unknown |
-0.104 |
0.006 |
| 35 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.104 |
-0.096 |