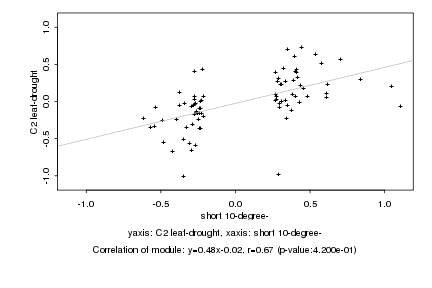
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
short 10-degree- |
Log2 signal ratio
C2 leaf-drought |
| 1 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.104 |
-0.056 |
| 2 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
1.042 |
0.207 |
| 3 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.837 |
0.302 |
| 4 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.698 |
0.576 |
| 5 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.616 |
0.230 |
| 6 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.606 |
0.059 |
| 7 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.606 |
0.109 |
| 8 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.575 |
0.516 |
| 9 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.532 |
0.644 |
| 10 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.482 |
0.072 |
| 11 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.454 |
0.182 |
| 12 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.442 |
0.730 |
| 13 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.431 |
0.216 |
| 14 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.428 |
-0.001 |
| 15 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.411 |
0.326 |
| 16 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.407 |
0.439 |
| 17 |
246125_at |
AT5G19875
|
unknown |
0.404 |
0.400 |
| 18 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.400 |
0.417 |
| 19 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.399 |
0.079 |
| 20 |
267036_at |
AT2G38465
|
unknown |
0.390 |
0.618 |
| 21 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.388 |
0.284 |
| 22 |
251753_at |
AT3G55760
|
unknown |
0.382 |
0.107 |
| 23 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.373 |
-0.120 |
| 24 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
0.346 |
0.709 |
| 25 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
0.345 |
-0.045 |
| 26 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
0.342 |
-0.224 |
| 27 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.334 |
0.281 |
| 28 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.333 |
0.025 |
| 29 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.317 |
0.448 |
| 30 |
258562_at |
AT3G05980
|
unknown |
0.304 |
0.006 |
| 31 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
0.303 |
0.231 |
| 32 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.301 |
0.235 |
| 33 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.294 |
-0.078 |
| 34 |
250151_at |
AT5G14570
|
ATNRT2.7, high affinity nitrate transporter 2.7, NRT2.7, high affinity nitrate transporter 2.7 |
0.292 |
-0.015 |
| 35 |
252127_at |
AT3G50960
|
PLP3a, phosducin-like protein 3 homolog |
0.287 |
0.312 |
| 36 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.283 |
-0.975 |
| 37 |
250053_at |
AT5G17850
|
[Sodium/calcium exchanger family protein] |
0.281 |
0.276 |
| 38 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
0.273 |
0.031 |
| 39 |
249091_at |
AT5G43860
|
ATCLH2, ARABIDOPSIS THALIANA CHLOROPHYLLASE 2, CLH2, chlorophyllase 2 |
0.271 |
0.080 |
| 40 |
249134_at |
AT5G43150
|
unknown |
0.268 |
0.104 |
| 41 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.266 |
0.016 |
| 42 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.265 |
0.404 |
| 43 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.618 |
-0.217 |
| 44 |
253305_at |
AT4G33666
|
unknown |
-0.575 |
-0.340 |
| 45 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.544 |
-0.329 |
| 46 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.537 |
-0.074 |
| 47 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.495 |
-0.249 |
| 48 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.486 |
-0.542 |
| 49 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.428 |
-0.670 |
| 50 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.400 |
-0.238 |
| 51 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.380 |
-0.044 |
| 52 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.378 |
0.125 |
| 53 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.353 |
-0.998 |
| 54 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.351 |
-0.502 |
| 55 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.346 |
-0.021 |
| 56 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.334 |
-0.337 |
| 57 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.312 |
-0.558 |
| 58 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.300 |
-0.655 |
| 59 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.300 |
-0.056 |
| 60 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.291 |
-0.303 |
| 61 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.282 |
-0.044 |
| 62 |
257880_at |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
-0.280 |
-0.038 |
| 63 |
259500_at |
AT1G15740
|
[Leucine-rich repeat family protein] |
-0.279 |
0.080 |
| 64 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.279 |
0.411 |
| 65 |
256569_at |
AT3G19550
|
unknown |
-0.278 |
-0.172 |
| 66 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.278 |
0.034 |
| 67 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.274 |
-0.585 |
| 68 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.269 |
-0.022 |
| 69 |
254851_at |
AT4G12010
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.266 |
-0.122 |
| 70 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.265 |
-0.147 |
| 71 |
248329_at |
AT5G52780
|
unknown |
-0.260 |
-0.148 |
| 72 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.248 |
-0.230 |
| 73 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.245 |
-0.153 |
| 74 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.243 |
-0.362 |
| 75 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.242 |
-0.091 |
| 76 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.237 |
0.001 |
| 77 |
259017_at |
AT3G07310
|
unknown |
-0.236 |
-0.092 |
| 78 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.235 |
-0.362 |
| 79 |
254705_at |
AT4G17870
|
PYR1, PYRABACTIN RESISTANCE 1, RCAR11, regulatory component of ABA receptor 11 |
-0.234 |
0.016 |
| 80 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.232 |
-0.160 |
| 81 |
257422_at |
AT1G11940
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.223 |
0.433 |
| 82 |
263875_at |
AT2G21970
|
SEP2, stress enhanced protein 2 |
-0.220 |
0.074 |
| 83 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.218 |
-0.191 |