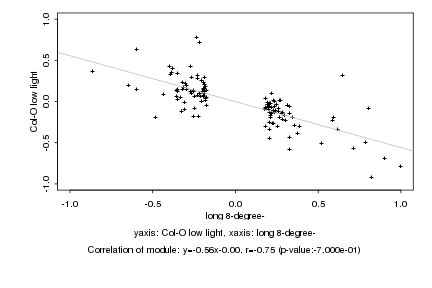
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
long 8-degree- |
Log2 signal ratio
Col-O low light |
| 1 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.995 |
-0.787 |
| 2 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.895 |
-0.689 |
| 3 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.819 |
-0.920 |
| 4 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.800 |
-0.081 |
| 5 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.784 |
-0.496 |
| 6 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.708 |
-0.569 |
| 7 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.644 |
0.319 |
| 8 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.615 |
-0.328 |
| 9 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.588 |
-0.183 |
| 10 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.581 |
-0.226 |
| 11 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.515 |
-0.498 |
| 12 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.382 |
-0.302 |
| 13 |
256924_at |
AT3G29590
|
AT5MAT |
0.374 |
-0.381 |
| 14 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.353 |
-0.285 |
| 15 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.339 |
-0.183 |
| 16 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.326 |
-0.572 |
| 17 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
0.326 |
-0.427 |
| 18 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
0.325 |
-0.142 |
| 19 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.324 |
-0.055 |
| 20 |
260903_at |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
0.313 |
-0.048 |
| 21 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.312 |
-0.038 |
| 22 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.297 |
-0.220 |
| 23 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.291 |
-0.160 |
| 24 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.284 |
-0.216 |
| 25 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.279 |
-0.127 |
| 26 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.277 |
-0.141 |
| 27 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.268 |
0.023 |
| 28 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.265 |
0.017 |
| 29 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.263 |
-0.193 |
| 30 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.258 |
-0.083 |
| 31 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
0.256 |
-0.110 |
| 32 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.249 |
-0.301 |
| 33 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
0.246 |
-0.031 |
| 34 |
262136_at |
AT1G77850
|
ARF17, auxin response factor 17 |
0.241 |
-0.097 |
| 35 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.235 |
0.007 |
| 36 |
259275_at |
AT3G01060
|
unknown |
0.232 |
-0.059 |
| 37 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
0.231 |
-0.130 |
| 38 |
254043_at |
AT4G25990
|
CIL |
0.227 |
0.013 |
| 39 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
0.225 |
-0.255 |
| 40 |
257252_at |
AT3G24170
|
ATGR1, glutathione-disulfide reductase, GR1, glutathione-disulfide reductase |
0.224 |
-0.103 |
| 41 |
257253_at |
AT3G24190
|
[Protein kinase superfamily protein] |
0.219 |
-0.259 |
| 42 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.215 |
-0.064 |
| 43 |
263805_at |
AT2G40400
|
unknown |
0.212 |
-0.122 |
| 44 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.212 |
0.104 |
| 45 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.212 |
-0.139 |
| 46 |
252412_at |
AT3G47295
|
unknown |
0.211 |
-0.183 |
| 47 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.210 |
-0.012 |
| 48 |
250758_at |
AT5G06000
|
ATEIF3G2, ARABIDOPSIS THALIANA EUKARYOTIC TRANSLATION INITIATION FACTOR 3G2, EIF3G2, eukaryotic translation initiation factor 3G2 |
0.208 |
-0.161 |
| 49 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.205 |
-0.438 |
| 50 |
245936_at |
AT5G19850
|
[alpha/beta-Hydrolases superfamily protein] |
0.204 |
-0.246 |
| 51 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.203 |
-0.336 |
| 52 |
246554_at |
AT5G15450
|
APG6, ALBINO AND PALE GREEN 6, AtCLPB3, CLPB-P, CASEIN LYTIC PROTEINASE B-P, CLPB3, casein lytic proteinase B3 |
0.203 |
-0.122 |
| 53 |
265768_at |
AT2G48020
|
[Major facilitator superfamily protein] |
0.202 |
-0.015 |
| 54 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.201 |
-0.060 |
| 55 |
264057_at |
AT2G28550
|
RAP2.7, related to AP2.7, TOE1, TARGET OF EARLY ACTIVATION TAGGED (EAT) 1 |
0.199 |
-0.089 |
| 56 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
0.198 |
-0.065 |
| 57 |
254054_at |
AT4G25320
|
AHL3, AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEIN 3 |
0.197 |
-0.054 |
| 58 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
0.195 |
-0.057 |
| 59 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
0.193 |
-0.034 |
| 60 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
0.188 |
-0.011 |
| 61 |
258056_at |
AT3G29010
|
[Biotin/lipoate A/B protein ligase family] |
0.179 |
-0.068 |
| 62 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
0.178 |
-0.082 |
| 63 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
0.177 |
-0.300 |
| 64 |
264941_at |
AT1G60680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.176 |
0.040 |
| 65 |
264824_at |
AT1G03420
|
Sadhu4-2, sadhu non-coding retrotransposon 4-2 |
0.175 |
-0.094 |
| 66 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.866 |
0.373 |
| 67 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
-0.647 |
0.201 |
| 68 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.601 |
0.158 |
| 69 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.598 |
0.634 |
| 70 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.489 |
-0.185 |
| 71 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.436 |
0.087 |
| 72 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.401 |
0.427 |
| 73 |
256096_at |
AT1G13650
|
unknown |
-0.396 |
0.340 |
| 74 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.390 |
0.359 |
| 75 |
258468_at |
AT3G06070
|
unknown |
-0.386 |
0.412 |
| 76 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.362 |
0.138 |
| 77 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.361 |
0.063 |
| 78 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.356 |
0.151 |
| 79 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.353 |
0.342 |
| 80 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.353 |
0.028 |
| 81 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.352 |
0.133 |
| 82 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
-0.334 |
0.050 |
| 83 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.327 |
-0.117 |
| 84 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.325 |
0.154 |
| 85 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.322 |
0.233 |
| 86 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.318 |
0.176 |
| 87 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.313 |
-0.094 |
| 88 |
262236_at |
AT1G48330
|
unknown |
-0.312 |
-0.004 |
| 89 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.307 |
0.228 |
| 90 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.300 |
0.198 |
| 91 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.300 |
0.155 |
| 92 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.275 |
0.128 |
| 93 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
-0.275 |
0.435 |
| 94 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.271 |
0.295 |
| 95 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.269 |
0.104 |
| 96 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.259 |
-0.170 |
| 97 |
256226_at |
AT1G56280
|
ATDI19, drought-induced 19, DI19, drought-induced 19 |
-0.257 |
0.126 |
| 98 |
262607_at |
AT1G13990
|
unknown |
-0.256 |
0.145 |
| 99 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
-0.251 |
-0.080 |
| 100 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.248 |
0.061 |
| 101 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.239 |
0.781 |
| 102 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.234 |
0.318 |
| 103 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.232 |
0.289 |
| 104 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.230 |
0.083 |
| 105 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.225 |
-0.175 |
| 106 |
266781_at |
AT2G28940
|
[Protein kinase superfamily protein] |
-0.223 |
0.100 |
| 107 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.223 |
0.107 |
| 108 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.218 |
0.721 |
| 109 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.215 |
0.071 |
| 110 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.209 |
0.263 |
| 111 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.208 |
0.005 |
| 112 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
-0.206 |
0.008 |
| 113 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.204 |
0.126 |
| 114 |
249072_at |
AT5G44060
|
unknown |
-0.199 |
0.149 |
| 115 |
246917_at |
AT5G25280
|
[serine-rich protein-related] |
-0.198 |
0.237 |
| 116 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.198 |
0.073 |
| 117 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.195 |
0.169 |
| 118 |
259924_at |
AT1G72740
|
[Homeodomain-like/winged-helix DNA-binding family protein] |
-0.195 |
0.079 |
| 119 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
-0.193 |
0.122 |
| 120 |
251704_at |
AT3G56360
|
unknown |
-0.193 |
0.300 |
| 121 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.193 |
0.211 |
| 122 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.192 |
0.160 |
| 123 |
248905_at |
AT5G46250
|
AtLARP6a, LARP6a, La related protein 6a |
-0.191 |
0.100 |
| 124 |
257868_at |
AT3G25070
|
RIN4, RPM1 interacting protein 4 |
-0.190 |
0.123 |
| 125 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.186 |
0.039 |
| 126 |
263668_at |
AT1G04350
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.186 |
0.023 |
| 127 |
266707_at |
AT2G03310
|
unknown |
-0.183 |
0.193 |
| 128 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.180 |
0.057 |
| 129 |
246012_at |
AT5G10650
|
[RING/U-box superfamily protein] |
-0.179 |
0.138 |
| 130 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.176 |
-0.044 |