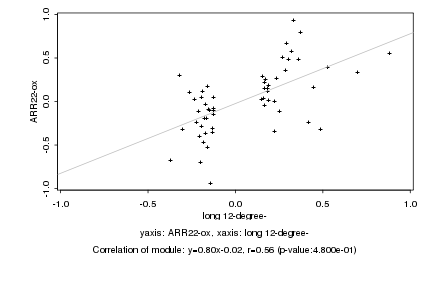
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
long 12-degree- |
Log2 signal ratio
ARR22-ox |
| 1 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.878 |
0.556 |
| 2 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.694 |
0.341 |
| 3 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.525 |
0.394 |
| 4 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.482 |
-0.320 |
| 5 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.446 |
0.167 |
| 6 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.417 |
-0.240 |
| 7 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.369 |
0.801 |
| 8 |
256924_at |
AT3G29590
|
AT5MAT |
0.356 |
0.491 |
| 9 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.331 |
0.942 |
| 10 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.316 |
0.586 |
| 11 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.303 |
0.491 |
| 12 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.292 |
0.667 |
| 13 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.286 |
0.363 |
| 14 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.266 |
0.516 |
| 15 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.251 |
-0.106 |
| 16 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.234 |
0.272 |
| 17 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.223 |
0.003 |
| 18 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.218 |
-0.339 |
| 19 |
259275_at |
AT3G01060
|
unknown |
0.189 |
0.018 |
| 20 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.185 |
0.191 |
| 21 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.181 |
0.159 |
| 22 |
257252_at |
AT3G24170
|
ATGR1, glutathione-disulfide reductase, GR1, glutathione-disulfide reductase |
0.178 |
0.120 |
| 23 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.169 |
0.262 |
| 24 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.163 |
-0.036 |
| 25 |
265954_at |
AT2G37260
|
ATWRKY44, DSL1, DR. STRANGELOVE 1, TTG2, TRANSPARENT TESTA GLABRA 2, WRKY44 |
0.162 |
0.229 |
| 26 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.162 |
0.159 |
| 27 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
0.160 |
0.045 |
| 28 |
246554_at |
AT5G15450
|
APG6, ALBINO AND PALE GREEN 6, AtCLPB3, CLPB-P, CASEIN LYTIC PROTEINASE B-P, CLPB3, casein lytic proteinase B3 |
0.150 |
0.291 |
| 29 |
245368_at |
AT4G15510
|
PPD1, PsbP-Domain Protein1 |
0.149 |
0.032 |
| 30 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.373 |
-0.677 |
| 31 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.321 |
0.302 |
| 32 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.307 |
-0.312 |
| 33 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
-0.266 |
0.113 |
| 34 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.239 |
0.030 |
| 35 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.225 |
-0.236 |
| 36 |
256096_at |
AT1G13650
|
unknown |
-0.212 |
-0.104 |
| 37 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.207 |
-0.396 |
| 38 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.205 |
-0.700 |
| 39 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.198 |
0.047 |
| 40 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.197 |
-0.279 |
| 41 |
254848_at |
AT4G11960
|
PGRL1B, PGR5-like B |
-0.189 |
0.118 |
| 42 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
-0.185 |
-0.467 |
| 43 |
254688_at |
AT4G13830
|
DJC26, DNA J protein C26, J20, DNAJ-like 20 |
-0.179 |
-0.187 |
| 44 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.177 |
-0.357 |
| 45 |
266005_at |
AT2G37340
|
AT-RS2Z33, arginine/serine-rich zinc knuckle-containing protein 33, ATRSZ33, ARGININE/SERINE-RICH ZINC KNUCKLE-CONTAINING PROTEIN 33, RS2Z33, arginine/serine-rich zinc knuckle-containing protein 33, RSZ33, arginine/serine-rich zinc knuckle-containing protein 33 |
-0.172 |
-0.028 |
| 46 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.171 |
-0.187 |
| 47 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.164 |
0.176 |
| 48 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.161 |
-0.518 |
| 49 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.158 |
-0.084 |
| 50 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.150 |
-0.099 |
| 51 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.143 |
-0.937 |
| 52 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.136 |
-0.301 |
| 53 |
260012_at |
AT1G67865
|
unknown |
-0.135 |
-0.353 |
| 54 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.131 |
-0.138 |
| 55 |
255957_at |
AT1G22160
|
unknown |
-0.130 |
0.049 |
| 56 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.129 |
-0.094 |
| 57 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.128 |
-0.077 |