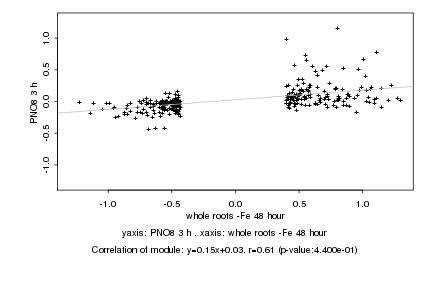
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots -Fe 48 hour |
Log2 signal ratio
PNO8 3 h |
| 1 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
1.294 |
0.025 |
| 2 |
262091_at |
AT1G56160
|
ATMYB72, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 72, MYB72, myb domain protein 72 |
1.268 |
0.056 |
| 3 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
1.223 |
0.263 |
| 4 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
1.197 |
0.031 |
| 5 |
263515_at |
AT2G21640
|
unknown |
1.141 |
0.220 |
| 6 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
1.141 |
-0.087 |
| 7 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
1.105 |
0.775 |
| 8 |
250152_at |
AT5G15120
|
unknown |
1.105 |
0.054 |
| 9 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.092 |
0.037 |
| 10 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.089 |
-0.016 |
| 11 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
1.063 |
0.233 |
| 12 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.055 |
0.192 |
| 13 |
246860_at |
AT5G25840
|
unknown |
1.052 |
-0.003 |
| 14 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
1.041 |
0.076 |
| 15 |
248028_at |
AT5G55620
|
unknown |
1.029 |
0.003 |
| 16 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.029 |
0.186 |
| 17 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
1.021 |
0.410 |
| 18 |
260522_x_at |
AT2G41730
|
unknown |
1.002 |
0.664 |
| 19 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.990 |
0.225 |
| 20 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.968 |
0.181 |
| 21 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.967 |
0.507 |
| 22 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.953 |
0.103 |
| 23 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.949 |
-0.165 |
| 24 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.930 |
0.054 |
| 25 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.895 |
0.085 |
| 26 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
0.894 |
-0.064 |
| 27 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.882 |
0.122 |
| 28 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.878 |
0.112 |
| 29 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.869 |
0.053 |
| 30 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.869 |
-0.052 |
| 31 |
254976_at |
AT4G10510
|
[Subtilase family protein] |
0.848 |
-0.054 |
| 32 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.844 |
0.001 |
| 33 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.843 |
0.526 |
| 34 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.840 |
0.204 |
| 35 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.813 |
0.062 |
| 36 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.805 |
0.063 |
| 37 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.805 |
0.023 |
| 38 |
264710_at |
AT1G09790
|
COBL6, COBRA-like protein 6 precursor |
0.805 |
0.038 |
| 39 |
246370_at |
AT1G51920
|
unknown |
0.803 |
0.089 |
| 40 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.801 |
1.168 |
| 41 |
251012_at |
AT5G02580
|
unknown |
0.795 |
0.031 |
| 42 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.793 |
0.217 |
| 43 |
258782_at |
AT3G11750
|
FOLB1 |
0.792 |
-0.079 |
| 44 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
0.779 |
0.195 |
| 45 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.779 |
0.207 |
| 46 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.772 |
0.009 |
| 47 |
260668_at |
AT1G19530
|
unknown |
0.769 |
-0.059 |
| 48 |
245076_at |
AT2G23170
|
GH3.3 |
0.736 |
0.294 |
| 49 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.728 |
0.043 |
| 50 |
264821_at |
AT1G03470
|
NET3A, Networked 3A |
0.726 |
0.032 |
| 51 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.719 |
0.082 |
| 52 |
261842_at |
AT1G16160
|
WAKL5, wall associated kinase-like 5 |
0.717 |
-0.085 |
| 53 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.717 |
0.116 |
| 54 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.714 |
0.563 |
| 55 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.706 |
-0.034 |
| 56 |
264635_at |
AT1G65500
|
unknown |
0.700 |
0.069 |
| 57 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.699 |
0.161 |
| 58 |
255075_at |
AT4G09110
|
[RING/U-box superfamily protein] |
0.693 |
0.032 |
| 59 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.684 |
0.497 |
| 60 |
256499_at |
AT1G36640
|
unknown |
0.668 |
-0.007 |
| 61 |
260754_at |
AT1G49000
|
unknown |
0.664 |
0.055 |
| 62 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
0.655 |
0.022 |
| 63 |
245401_at |
AT4G17670
|
unknown |
0.651 |
0.097 |
| 64 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.643 |
0.425 |
| 65 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.639 |
0.234 |
| 66 |
254571_at |
AT4G19370
|
unknown |
0.638 |
-0.039 |
| 67 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.628 |
-0.023 |
| 68 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
0.627 |
-0.039 |
| 69 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.623 |
0.478 |
| 70 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
0.623 |
-0.024 |
| 71 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.601 |
0.553 |
| 72 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.589 |
0.064 |
| 73 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.586 |
0.263 |
| 74 |
253796_at |
AT4G28460
|
unknown |
0.583 |
0.121 |
| 75 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.580 |
0.226 |
| 76 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.579 |
0.262 |
| 77 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
0.579 |
-0.061 |
| 78 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.572 |
0.179 |
| 79 |
251293_at |
AT3G61930
|
unknown |
0.571 |
0.168 |
| 80 |
258029_at |
AT3G27580
|
ATPK7, D6PKL3, D6 PROTEIN KINASE LIKE 3 |
0.569 |
0.096 |
| 81 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.558 |
0.060 |
| 82 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.553 |
0.660 |
| 83 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.551 |
0.191 |
| 84 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.550 |
0.736 |
| 85 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.547 |
-0.057 |
| 86 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.542 |
0.093 |
| 87 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.541 |
0.064 |
| 88 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.539 |
0.032 |
| 89 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.530 |
0.295 |
| 90 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.528 |
0.169 |
| 91 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.526 |
0.362 |
| 92 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.519 |
0.181 |
| 93 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.516 |
-0.046 |
| 94 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
0.516 |
0.198 |
| 95 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.514 |
0.041 |
| 96 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.511 |
0.142 |
| 97 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.503 |
0.174 |
| 98 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.502 |
0.113 |
| 99 |
258203_at |
AT3G13950
|
unknown |
0.502 |
0.099 |
| 100 |
245208_at |
AT5G12330
|
LRP1, LATERAL ROOT PRIMORDIUM 1 |
0.494 |
0.096 |
| 101 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.490 |
0.350 |
| 102 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.488 |
0.036 |
| 103 |
267063_at |
AT2G41120
|
unknown |
0.487 |
0.059 |
| 104 |
260849_at |
AT1G21860
|
sks7, SKU5 similar 7 |
0.482 |
-0.027 |
| 105 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.482 |
0.050 |
| 106 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.482 |
0.255 |
| 107 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.481 |
0.110 |
| 108 |
264680_at |
AT1G65510
|
unknown |
0.478 |
-0.140 |
| 109 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.472 |
0.126 |
| 110 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
0.472 |
0.017 |
| 111 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.470 |
0.079 |
| 112 |
267572_at |
AT2G30660
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.468 |
-0.034 |
| 113 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.459 |
-0.063 |
| 114 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.459 |
0.096 |
| 115 |
249636_at |
AT5G36890
|
BGLU42, beta glucosidase 42 |
0.458 |
-0.020 |
| 116 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.457 |
0.584 |
| 117 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
0.456 |
0.075 |
| 118 |
263985_at |
AT2G42750
|
DJC77, DNA J protein C77 |
0.452 |
0.198 |
| 119 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
0.451 |
0.085 |
| 120 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.450 |
0.022 |
| 121 |
254020_at |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
0.448 |
0.036 |
| 122 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.446 |
0.057 |
| 123 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.441 |
0.145 |
| 124 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.433 |
0.087 |
| 125 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.430 |
0.045 |
| 126 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.429 |
-0.026 |
| 127 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.429 |
0.074 |
| 128 |
253305_at |
AT4G33666
|
unknown |
0.427 |
-0.012 |
| 129 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.424 |
-0.020 |
| 130 |
260472_at |
AT1G10990
|
unknown |
0.423 |
-0.028 |
| 131 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.422 |
0.128 |
| 132 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.421 |
-0.107 |
| 133 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.416 |
0.065 |
| 134 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
0.414 |
0.095 |
| 135 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.414 |
0.033 |
| 136 |
245423_at |
AT4G17483
|
[alpha/beta-Hydrolases superfamily protein] |
0.412 |
-0.053 |
| 137 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.411 |
0.264 |
| 138 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
0.411 |
-0.094 |
| 139 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.407 |
0.039 |
| 140 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.403 |
0.078 |
| 141 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.403 |
0.110 |
| 142 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.401 |
-0.031 |
| 143 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.401 |
0.985 |
| 144 |
253179_at |
AT4G35200
|
unknown |
0.397 |
-0.019 |
| 145 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
0.397 |
-0.028 |
| 146 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.397 |
0.239 |
| 147 |
264082_at |
AT2G28570
|
unknown |
0.395 |
0.019 |
| 148 |
260495_at |
AT2G41810
|
unknown |
-1.228 |
-0.006 |
| 149 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-1.146 |
-0.174 |
| 150 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-1.116 |
-0.025 |
| 151 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-1.046 |
-0.114 |
| 152 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-1.019 |
-0.024 |
| 153 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.990 |
-0.028 |
| 154 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.962 |
-0.095 |
| 155 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.958 |
-0.091 |
| 156 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.947 |
-0.240 |
| 157 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.920 |
-0.228 |
| 158 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.873 |
-0.159 |
| 159 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-0.873 |
-0.192 |
| 160 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.860 |
-0.107 |
| 161 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.855 |
-0.190 |
| 162 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.851 |
-0.059 |
| 163 |
247320_at |
AT5G64040
|
PSAN |
-0.826 |
-0.019 |
| 164 |
263840_at |
AT2G36885
|
unknown |
-0.825 |
-0.147 |
| 165 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.788 |
-0.253 |
| 166 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
-0.777 |
-0.172 |
| 167 |
249358_at |
AT5G40510
|
[Sucrase/ferredoxin-like family protein] |
-0.772 |
-0.083 |
| 168 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
-0.761 |
0.008 |
| 169 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.747 |
-0.165 |
| 170 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.745 |
-0.027 |
| 171 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.736 |
-0.188 |
| 172 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
-0.728 |
0.025 |
| 173 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.727 |
-0.124 |
| 174 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.705 |
-0.032 |
| 175 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.705 |
-0.008 |
| 176 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.705 |
-0.160 |
| 177 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
-0.701 |
0.062 |
| 178 |
262277_at |
AT1G68650
|
[Uncharacterized protein family (UPF0016)] |
-0.697 |
-0.216 |
| 179 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
-0.689 |
-0.018 |
| 180 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.684 |
-0.432 |
| 181 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.675 |
0.022 |
| 182 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.675 |
-0.019 |
| 183 |
254187_at |
AT4G23890
|
CRR31, CHLORORESPIRATORY REDUCTION 31, NdhS, NADH dehydrogenase-like complex S |
-0.670 |
-0.021 |
| 184 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.670 |
-0.094 |
| 185 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.656 |
-0.239 |
| 186 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
-0.650 |
-0.073 |
| 187 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
-0.645 |
-0.130 |
| 188 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
-0.645 |
-0.034 |
| 189 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.641 |
-0.174 |
| 190 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.640 |
-0.058 |
| 191 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.632 |
-0.421 |
| 192 |
262954_at |
AT1G54500
|
[Rubredoxin-like superfamily protein] |
-0.625 |
-0.029 |
| 193 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.623 |
0.013 |
| 194 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.604 |
-0.158 |
| 195 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.603 |
-0.080 |
| 196 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
-0.599 |
-0.031 |
| 197 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.598 |
-0.024 |
| 198 |
260337_at |
AT1G69310
|
ATWRKY57, WRKY57, WRKY DNA-binding protein 57 |
-0.591 |
-0.010 |
| 199 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.591 |
-0.009 |
| 200 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.590 |
-0.223 |
| 201 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
-0.586 |
-0.092 |
| 202 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.582 |
-0.049 |
| 203 |
253393_at |
AT4G32690
|
ATGLB3, ARABIDOPSIS HEMOGLOBIN 3, GLB3, hemoglobin 3 |
-0.582 |
-0.043 |
| 204 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.581 |
-0.043 |
| 205 |
245195_at |
AT1G67740
|
PSBY, photosystem II BY, YCF32 |
-0.579 |
0.012 |
| 206 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.579 |
0.003 |
| 207 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.578 |
-0.105 |
| 208 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-0.577 |
0.028 |
| 209 |
251466_at |
AT3G59340
|
unknown |
-0.577 |
-0.180 |
| 210 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.577 |
-0.116 |
| 211 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.573 |
-0.063 |
| 212 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.572 |
-0.023 |
| 213 |
257311_at |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
-0.563 |
-0.018 |
| 214 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.562 |
-0.002 |
| 215 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.562 |
-0.137 |
| 216 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.561 |
-0.088 |
| 217 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.560 |
-0.420 |
| 218 |
251116_at |
AT3G63470
|
scpl40, serine carboxypeptidase-like 40 |
-0.560 |
-0.047 |
| 219 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.553 |
0.012 |
| 220 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.552 |
-0.024 |
| 221 |
264826_at |
AT1G03410
|
2A6 |
-0.551 |
-0.119 |
| 222 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.551 |
0.139 |
| 223 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
-0.551 |
-0.016 |
| 224 |
259896_at |
AT1G71500
|
[Rieske (2Fe-2S) domain-containing protein] |
-0.548 |
0.013 |
| 225 |
245061_at |
AT2G39730
|
RCA, rubisco activase |
-0.543 |
-0.001 |
| 226 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
-0.542 |
0.009 |
| 227 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.540 |
-0.028 |
| 228 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.539 |
-0.042 |
| 229 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
-0.538 |
0.005 |
| 230 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.536 |
-0.138 |
| 231 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.532 |
-0.022 |
| 232 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.527 |
0.076 |
| 233 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.526 |
-0.204 |
| 234 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.523 |
0.131 |
| 235 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.519 |
-0.097 |
| 236 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.513 |
0.004 |
| 237 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.513 |
0.015 |
| 238 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
-0.511 |
-0.002 |
| 239 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.508 |
-0.011 |
| 240 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
-0.506 |
-0.121 |
| 241 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.502 |
-0.073 |
| 242 |
260236_at |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
-0.500 |
-0.019 |
| 243 |
256283_at |
AT3G12540
|
unknown |
-0.500 |
-0.056 |
| 244 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.499 |
-0.060 |
| 245 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.497 |
-0.112 |
| 246 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
-0.495 |
0.019 |
| 247 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.493 |
-0.097 |
| 248 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.487 |
-0.138 |
| 249 |
263736_at |
AT1G60000
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.487 |
0.004 |
| 250 |
247476_at |
AT5G62330
|
unknown |
-0.485 |
-0.078 |
| 251 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.480 |
-0.035 |
| 252 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.479 |
-0.018 |
| 253 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
-0.478 |
-0.006 |
| 254 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
-0.477 |
0.056 |
| 255 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.477 |
0.017 |
| 256 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.477 |
-0.183 |
| 257 |
260161_at |
AT1G79860
|
ATROPGEF12, MEE64, MATERNAL EFFECT EMBRYO ARREST 64, ROPGEF12, ROP (rho of plants) guanine nucleotide exchange factor 12 |
-0.475 |
0.061 |
| 258 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.474 |
-0.142 |
| 259 |
246001_at |
AT5G20790
|
unknown |
-0.472 |
0.119 |
| 260 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.472 |
0.036 |
| 261 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.471 |
-0.002 |
| 262 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.471 |
-0.046 |
| 263 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.470 |
-0.048 |
| 264 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.470 |
-0.090 |
| 265 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.469 |
-0.053 |
| 266 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.469 |
-0.135 |
| 267 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.468 |
-0.114 |
| 268 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
-0.467 |
0.044 |
| 269 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
-0.466 |
0.012 |
| 270 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.464 |
-0.076 |
| 271 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
-0.461 |
0.015 |
| 272 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
-0.460 |
0.168 |
| 273 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.459 |
-0.169 |
| 274 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.455 |
-0.188 |
| 275 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.455 |
-0.194 |
| 276 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.454 |
-0.026 |
| 277 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.454 |
0.098 |
| 278 |
260014_at |
AT1G68010
|
ATHPR1, HPR, hydroxypyruvate reductase |
-0.453 |
0.011 |
| 279 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.451 |
-0.143 |
| 280 |
247131_at |
AT5G66190
|
ATLFNR1, LEAF FNR 1, FNR1, ferredoxin-NADP(+)-oxidoreductase 1, LFNR1, leaf-type chloroplast-targeted FNR 1 |
-0.451 |
0.020 |
| 281 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
-0.451 |
0.067 |
| 282 |
251501_at |
AT3G59120
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.450 |
-0.010 |
| 283 |
247722_at |
AT5G59150
|
ATRAB-A2D, ARABIDOPSIS RAB GTPASE HOMOLOG A2D, ATRABA2D, RAB GTPase homolog A2D, RABA2D, RAB GTPase homolog A2D |
-0.449 |
-0.074 |
| 284 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.448 |
-0.051 |
| 285 |
246617_at |
AT5G36270
|
[similar to DHAR2, glutathione dehydrogenase (ascorbate) [Arabidopsis thaliana] (TAIR:AT1G75270.1)] |
-0.446 |
-0.019 |
| 286 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.445 |
-0.067 |
| 287 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.442 |
0.009 |
| 288 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.442 |
-0.115 |
| 289 |
245510_at |
AT4G15740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.442 |
-0.040 |
| 290 |
255142_at |
AT4G08390
|
SAPX, stromal ascorbate peroxidase |
-0.440 |
-0.087 |
| 291 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.438 |
-0.084 |
| 292 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.438 |
0.008 |
| 293 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.437 |
-0.231 |