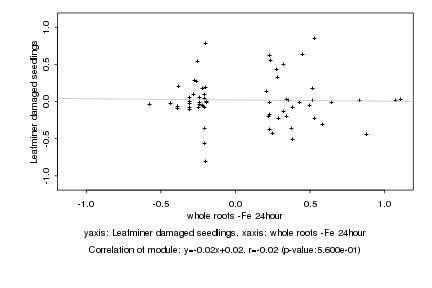
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots -Fe 24hour |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
262091_at |
AT1G56160
|
ATMYB72, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 72, MYB72, myb domain protein 72 |
1.104 |
0.033 |
| 2 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
1.072 |
0.022 |
| 3 |
246860_at |
AT5G25840
|
unknown |
0.878 |
-0.442 |
| 4 |
257135_at |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.827 |
0.016 |
| 5 |
257689_at |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
0.644 |
-0.010 |
| 6 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.583 |
-0.308 |
| 7 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.528 |
0.852 |
| 8 |
264821_at |
AT1G03470
|
NET3A, Networked 3A |
0.526 |
-0.216 |
| 9 |
264710_at |
AT1G09790
|
COBL6, COBRA-like protein 6 precursor |
0.516 |
0.015 |
| 10 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.511 |
0.186 |
| 11 |
250152_at |
AT5G15120
|
unknown |
0.490 |
-0.041 |
| 12 |
251293_at |
AT3G61930
|
unknown |
0.443 |
0.635 |
| 13 |
254976_at |
AT4G10510
|
[Subtilase family protein] |
0.426 |
-0.004 |
| 14 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.381 |
-0.077 |
| 15 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.377 |
-0.503 |
| 16 |
254020_at |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
0.370 |
-0.359 |
| 17 |
252145_at |
AT3G51200
|
SAUR18, SMALL AUXIN UPREGULATED RNA 18 |
0.353 |
0.026 |
| 18 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.342 |
0.039 |
| 19 |
263985_at |
AT2G42750
|
DJC77, DNA J protein C77 |
0.338 |
-0.194 |
| 20 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.322 |
0.505 |
| 21 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
0.320 |
-0.123 |
| 22 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
0.287 |
-0.222 |
| 23 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.279 |
0.329 |
| 24 |
264680_at |
AT1G65510
|
unknown |
0.270 |
0.432 |
| 25 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
0.246 |
-0.424 |
| 26 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.230 |
0.563 |
| 27 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.228 |
-0.372 |
| 28 |
247570_at |
AT5G61250
|
AtGUS1, glucuronidase 1, GUS1, glucuronidase 1 |
0.223 |
-0.012 |
| 29 |
245401_at |
AT4G17670
|
unknown |
0.223 |
-0.171 |
| 30 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.223 |
0.629 |
| 31 |
247001_at |
AT5G67330
|
ATNRAMP4, ARABIDOPSIS THALIANA NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN 4, NRAMP4, natural resistance associated macrophage protein 4 |
0.217 |
-0.199 |
| 32 |
246614_at |
AT5G35410
|
ATSOS2, SALT OVERLY SENSITIVE 2, CIPK24, CBL-INTERACTING PROTEIN KINASE 24, SNRK3.11, SNF1-RELATED PROTEIN KINASE 3.11, SOS2, SALT OVERLY SENSITIVE 2 |
0.202 |
0.137 |
| 33 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.581 |
-0.028 |
| 34 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
-0.442 |
-0.019 |
| 35 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.394 |
-0.057 |
| 36 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.392 |
-0.087 |
| 37 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.385 |
0.204 |
| 38 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.314 |
0.001 |
| 39 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.312 |
-0.104 |
| 40 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
-0.311 |
0.064 |
| 41 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.310 |
-0.026 |
| 42 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.310 |
-0.075 |
| 43 |
249358_at |
AT5G40510
|
[Sucrase/ferredoxin-like family protein] |
-0.285 |
0.103 |
| 44 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.281 |
0.294 |
| 45 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
-0.262 |
0.280 |
| 46 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.259 |
0.543 |
| 47 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.254 |
-0.068 |
| 48 |
249566_at |
AT5G38450
|
CYP735A1, cytochrome P450, family 735, subfamily A, polypeptide 1 |
-0.247 |
0.063 |
| 49 |
251863_at |
AT3G54870
|
ARK1, ARMADILLO REPEAT-CONTAINING KINESIN 1, AtKINUc, Arabidopsis thaliana KINESIN Ungrouped clade, gene A, CAE1, CA-ROP2 ENHANCER 1, MRH2, MORPHOGENESIS OF ROOT HAIR 2 |
-0.243 |
-0.027 |
| 50 |
252510_at |
AT3G46270
|
[receptor protein kinase-related] |
-0.242 |
-0.003 |
| 51 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.225 |
0.187 |
| 52 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
-0.222 |
-0.052 |
| 53 |
259028_at |
AT3G09290
|
TAC1, telomerase activator1 |
-0.216 |
-0.063 |
| 54 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.214 |
-0.559 |
| 55 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.211 |
-0.353 |
| 56 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.211 |
-0.080 |
| 57 |
245946_at |
AT5G19580
|
[glyoxal oxidase-related protein] |
-0.209 |
0.108 |
| 58 |
260433_at |
AT1G68170
|
UMAMIT23, Usually multiple acids move in and out Transporters 23 |
-0.208 |
0.054 |
| 59 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.205 |
0.197 |
| 60 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.203 |
-0.797 |
| 61 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.201 |
0.792 |
| 62 |
256283_at |
AT3G12540
|
unknown |
-0.197 |
0.002 |
| 63 |
253354_at |
AT4G33370
|
[DEA(D/H)-box RNA helicase family protein] |
-0.197 |
-0.001 |