|
probeID |
AGICode |
Annotation |
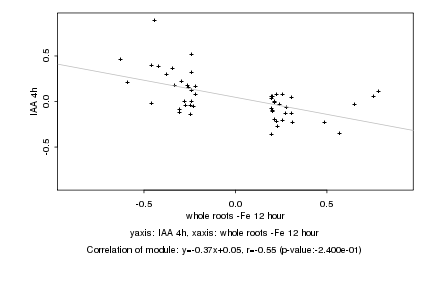
Log2 signal ratio
whole roots -Fe 12 hour |
Log2 signal ratio
IAA 4h |
| 1 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.779 |
0.114 |
| 2 |
253502_at |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
0.751 |
0.056 |
| 3 |
262091_at |
AT1G56160
|
ATMYB72, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 72, MYB72, myb domain protein 72 |
0.650 |
-0.024 |
| 4 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.564 |
-0.345 |
| 5 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.486 |
-0.225 |
| 6 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.311 |
-0.223 |
| 7 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.305 |
-0.121 |
| 8 |
249557_at |
AT5G38280
|
PR5K, PR5-like receptor kinase |
0.303 |
0.048 |
| 9 |
258604_at |
AT3G02980
|
MCC1, MEIOTIC CONTROL OF CROSSOVERS1 |
0.275 |
-0.063 |
| 10 |
264054_at |
AT2G22540
|
AGL22, AGAMOUS-like 22, FAQ1, Flowering Arabidopsis QTL1, SVP, SHORT VEGETATIVE PHASE |
0.273 |
-0.128 |
| 11 |
264759_at |
AT1G61480
|
[S-locus lectin protein kinase family protein] |
0.257 |
0.081 |
| 12 |
251243_at |
AT3G61870
|
unknown |
0.253 |
-0.208 |
| 13 |
260210_at |
AT1G74420
|
ATFUT3, FUT3, fucosyltransferase 3 |
0.236 |
-0.027 |
| 14 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
0.226 |
-0.272 |
| 15 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
0.223 |
-0.209 |
| 16 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.219 |
0.085 |
| 17 |
252907_at |
AT4G39650
|
GGT2, gamma-glutamyl transpeptidase 2 |
0.210 |
0.001 |
| 18 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
0.210 |
-0.192 |
| 19 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.210 |
0.000 |
| 20 |
254014_at |
AT4G26120
|
[Ankyrin repeat family protein / BTB/POZ domain-containing protein] |
0.201 |
-0.104 |
| 21 |
247875_at |
AT5G57720
|
[AP2/B3-like transcriptional factor family protein] |
0.199 |
-0.088 |
| 22 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
0.198 |
0.059 |
| 23 |
254807_at |
AT4G12700
|
unknown |
0.197 |
0.059 |
| 24 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.197 |
-0.358 |
| 25 |
250345_at |
AT5G11940
|
[Subtilase family protein] |
0.193 |
-0.074 |
| 26 |
246548_at |
AT5G14910
|
[Heavy metal transport/detoxification superfamily protein ] |
0.193 |
0.038 |
| 27 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
-0.629 |
0.469 |
| 28 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.592 |
0.209 |
| 29 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.462 |
0.400 |
| 30 |
260495_at |
AT2G41810
|
unknown |
-0.461 |
-0.021 |
| 31 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.443 |
0.900 |
| 32 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
-0.422 |
0.388 |
| 33 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.377 |
0.305 |
| 34 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.349 |
0.367 |
| 35 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.334 |
0.176 |
| 36 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
-0.311 |
-0.085 |
| 37 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.310 |
-0.113 |
| 38 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.296 |
0.226 |
| 39 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.279 |
0.003 |
| 40 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
-0.275 |
-0.040 |
| 41 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.265 |
0.184 |
| 42 |
245510_at |
AT4G15740
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.259 |
0.158 |
| 43 |
264129_at |
AT1G79170
|
unknown |
-0.250 |
-0.039 |
| 44 |
253296_at |
AT4G33770
|
ITPK2, inositol 1,3,4-trisphosphate 5/6 kinase 2 |
-0.248 |
-0.139 |
| 45 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
-0.244 |
0.004 |
| 46 |
256506_at |
AT1G75160
|
unknown |
-0.244 |
0.125 |
| 47 |
246807_at |
AT5G27100
|
ATGLR2.1, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 2.1, GLR2.1, glutamate receptor 2.1 |
-0.243 |
0.523 |
| 48 |
252205_at |
AT3G50350
|
unknown |
-0.243 |
0.323 |
| 49 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
-0.230 |
-0.049 |
| 50 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.222 |
0.080 |
| 51 |
254092_at |
AT4G25090
|
[Riboflavin synthase-like superfamily protein] |
-0.220 |
0.165 |