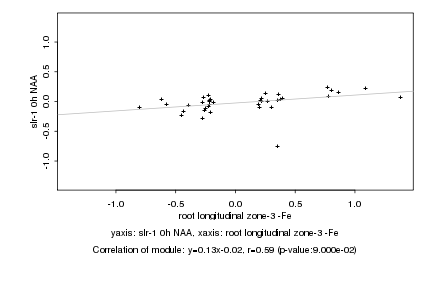
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root longitudinal zone-3 -Fe |
Log2 signal ratio
slr-1 0h NAA |
| 1 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
1.378 |
0.084 |
| 2 |
245692_at |
AT5G04150
|
BHLH101 |
1.088 |
0.235 |
| 3 |
262373_at |
AT1G73120
|
unknown |
0.857 |
0.168 |
| 4 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.801 |
0.188 |
| 5 |
245296_at |
AT4G16370
|
oligopeptide transporter |
0.771 |
0.087 |
| 6 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.764 |
0.249 |
| 7 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.391 |
0.054 |
| 8 |
264127_at |
AT1G79250
|
AGC1.7, AGC kinase 1.7 |
0.373 |
0.044 |
| 9 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
0.358 |
0.131 |
| 10 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.351 |
-0.746 |
| 11 |
260797_at |
AT1G78390
|
ATNCED9, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9, NCED9, nine-cis-epoxycarotenoid dioxygenase 9 |
0.347 |
0.030 |
| 12 |
263465_at |
AT2G31940
|
unknown |
0.301 |
-0.094 |
| 13 |
263112_at |
AT1G03080
|
NET1D, Networked 1D |
0.268 |
0.008 |
| 14 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.249 |
0.143 |
| 15 |
257421_at |
AT1G12030
|
unknown |
0.213 |
0.062 |
| 16 |
248218_at |
AT5G53710
|
unknown |
0.211 |
0.016 |
| 17 |
263002_at |
AT1G54200
|
unknown |
0.203 |
0.027 |
| 18 |
246151_at |
AT5G19950
|
unknown |
0.196 |
-0.087 |
| 19 |
265828_at |
AT2G14520
|
unknown |
0.192 |
-0.040 |
| 20 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.809 |
-0.095 |
| 21 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-0.626 |
0.040 |
| 22 |
263831_at |
AT2G40300
|
ATFER4, ferritin 4, FER4, ferritin 4 |
-0.583 |
-0.034 |
| 23 |
247320_at |
AT5G64040
|
PSAN |
-0.454 |
-0.221 |
| 24 |
263840_at |
AT2G36885
|
unknown |
-0.440 |
-0.156 |
| 25 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.399 |
-0.059 |
| 26 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.283 |
-0.280 |
| 27 |
255142_at |
AT4G08390
|
SAPX, stromal ascorbate peroxidase |
-0.279 |
-0.011 |
| 28 |
250270_at |
AT5G12980
|
[Cell differentiation, Rcd1-like protein] |
-0.270 |
0.077 |
| 29 |
246415_at |
AT5G17160
|
unknown |
-0.265 |
-0.144 |
| 30 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
-0.251 |
-0.116 |
| 31 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
-0.233 |
-0.069 |
| 32 |
256449_at |
AT1G33460
|
[Mutator-like transposase family, has a 3.6e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.228 |
0.106 |
| 33 |
265025_at |
AT1G24575
|
unknown |
-0.225 |
-0.045 |
| 34 |
257589_at |
AT1G55050
|
unknown |
-0.222 |
0.011 |
| 35 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.221 |
0.025 |
| 36 |
248339_at |
AT5G52520
|
OVA6, OVULE ABORTION 6, PRORS1, PROLYL-TRNA SYNTHETASE 1 |
-0.211 |
0.047 |
| 37 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
-0.211 |
-0.182 |
| 38 |
251909_at |
AT3G53790
|
TRFL4, TRF-like 4 |
-0.191 |
-0.005 |