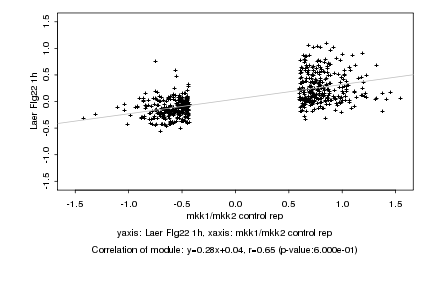
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
mkk1/mkk2 control rep |
Log2 signal ratio
Laer Flg22 1h |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.542 |
0.061 |
| 2 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.449 |
0.185 |
| 3 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.414 |
0.042 |
| 4 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
1.372 |
-0.186 |
| 5 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
1.370 |
0.160 |
| 6 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.319 |
0.068 |
| 7 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
1.312 |
0.688 |
| 8 |
245076_at |
AT2G23170
|
GH3.3 |
1.306 |
0.046 |
| 9 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
1.223 |
0.502 |
| 10 |
267345_at |
AT2G44240
|
unknown |
1.221 |
0.086 |
| 11 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.215 |
0.163 |
| 12 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
1.204 |
0.106 |
| 13 |
258203_at |
AT3G13950
|
unknown |
1.197 |
0.370 |
| 14 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.187 |
0.911 |
| 15 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.184 |
0.125 |
| 16 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
1.183 |
0.254 |
| 17 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
1.179 |
0.460 |
| 18 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
1.179 |
0.125 |
| 19 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
1.143 |
0.433 |
| 20 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
1.130 |
0.082 |
| 21 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.117 |
0.693 |
| 22 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.114 |
0.175 |
| 23 |
259385_at |
AT1G13470
|
unknown |
1.107 |
0.197 |
| 24 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
1.106 |
-0.075 |
| 25 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
1.096 |
0.879 |
| 26 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.083 |
-0.129 |
| 27 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
1.074 |
0.592 |
| 28 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
1.067 |
-0.007 |
| 29 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
1.063 |
0.026 |
| 30 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
1.060 |
0.650 |
| 31 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
1.058 |
0.310 |
| 32 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
1.045 |
0.315 |
| 33 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
1.037 |
0.383 |
| 34 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
1.036 |
0.283 |
| 35 |
264680_at |
AT1G65510
|
unknown |
1.036 |
0.201 |
| 36 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
1.034 |
0.242 |
| 37 |
249004_at |
AT5G44570
|
unknown |
1.029 |
0.102 |
| 38 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
1.029 |
0.026 |
| 39 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.024 |
0.108 |
| 40 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
1.020 |
0.524 |
| 41 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
1.020 |
0.498 |
| 42 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.014 |
0.593 |
| 43 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
1.014 |
0.294 |
| 44 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
1.009 |
0.429 |
| 45 |
267178_at |
AT2G37750
|
unknown |
1.003 |
0.157 |
| 46 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
1.002 |
0.279 |
| 47 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
1.000 |
0.028 |
| 48 |
251419_at |
AT3G60470
|
unknown |
0.999 |
0.202 |
| 49 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.998 |
0.903 |
| 50 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.993 |
0.456 |
| 51 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.993 |
-0.203 |
| 52 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.985 |
-0.003 |
| 53 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.984 |
0.082 |
| 54 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.983 |
0.780 |
| 55 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.982 |
-0.034 |
| 56 |
264580_at |
AT1G05340
|
unknown |
0.980 |
0.366 |
| 57 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.979 |
0.067 |
| 58 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.970 |
0.274 |
| 59 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.966 |
-0.002 |
| 60 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.962 |
0.103 |
| 61 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.961 |
0.516 |
| 62 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.953 |
-0.059 |
| 63 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.943 |
0.812 |
| 64 |
249454_at |
AT5G39520
|
unknown |
0.939 |
-0.029 |
| 65 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.932 |
0.043 |
| 66 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.931 |
0.063 |
| 67 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.930 |
-0.155 |
| 68 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.927 |
0.541 |
| 69 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.922 |
0.066 |
| 70 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.922 |
0.213 |
| 71 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.917 |
0.098 |
| 72 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.910 |
1.032 |
| 73 |
260668_at |
AT1G19530
|
unknown |
0.896 |
0.020 |
| 74 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.890 |
0.344 |
| 75 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.889 |
0.493 |
| 76 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.889 |
0.974 |
| 77 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.883 |
0.035 |
| 78 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.881 |
0.711 |
| 79 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
0.880 |
-0.042 |
| 80 |
251633_at |
AT3G57460
|
[catalytics] |
0.879 |
0.356 |
| 81 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.876 |
0.155 |
| 82 |
253827_at |
AT4G28085
|
unknown |
0.873 |
0.704 |
| 83 |
251612_at |
AT3G57950
|
unknown |
0.872 |
0.206 |
| 84 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.871 |
0.716 |
| 85 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.871 |
0.237 |
| 86 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.869 |
0.038 |
| 87 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.869 |
0.130 |
| 88 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.865 |
0.502 |
| 89 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.859 |
0.630 |
| 90 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.857 |
0.062 |
| 91 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.857 |
0.240 |
| 92 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.855 |
0.663 |
| 93 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.855 |
0.086 |
| 94 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.855 |
0.546 |
| 95 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.853 |
0.075 |
| 96 |
252345_at |
AT3G48640
|
unknown |
0.851 |
0.313 |
| 97 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.851 |
1.100 |
| 98 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.849 |
0.329 |
| 99 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.847 |
0.440 |
| 100 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.846 |
-0.046 |
| 101 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.841 |
0.038 |
| 102 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.837 |
0.794 |
| 103 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
0.837 |
0.006 |
| 104 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.834 |
0.415 |
| 105 |
258209_at |
AT3G14060
|
unknown |
0.834 |
-0.302 |
| 106 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.833 |
0.154 |
| 107 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
0.831 |
0.743 |
| 108 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.831 |
0.548 |
| 109 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.828 |
0.225 |
| 110 |
259489_at |
AT1G15790
|
unknown |
0.826 |
0.119 |
| 111 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.823 |
0.572 |
| 112 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.823 |
0.265 |
| 113 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.821 |
0.262 |
| 114 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.818 |
0.099 |
| 115 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.817 |
-0.122 |
| 116 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.815 |
0.122 |
| 117 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.814 |
0.163 |
| 118 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.813 |
0.143 |
| 119 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.809 |
-0.127 |
| 120 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.808 |
0.363 |
| 121 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.804 |
0.571 |
| 122 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.804 |
0.568 |
| 123 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.802 |
0.074 |
| 124 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.799 |
0.616 |
| 125 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.798 |
0.000 |
| 126 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.797 |
0.260 |
| 127 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.795 |
0.434 |
| 128 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.795 |
0.656 |
| 129 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.795 |
0.075 |
| 130 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.793 |
1.021 |
| 131 |
263475_at |
AT2G31945
|
unknown |
0.792 |
0.766 |
| 132 |
260744_at |
AT1G15010
|
unknown |
0.791 |
0.410 |
| 133 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.790 |
-0.077 |
| 134 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.789 |
-0.000 |
| 135 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.789 |
0.304 |
| 136 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
0.787 |
0.161 |
| 137 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.787 |
0.370 |
| 138 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.784 |
0.562 |
| 139 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.783 |
0.518 |
| 140 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.782 |
0.332 |
| 141 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.780 |
0.194 |
| 142 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.778 |
0.120 |
| 143 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.777 |
0.291 |
| 144 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.775 |
-0.115 |
| 145 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.775 |
0.694 |
| 146 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.775 |
0.239 |
| 147 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.774 |
0.776 |
| 148 |
255912_at |
AT1G66960
|
LUP5 |
0.774 |
0.040 |
| 149 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.773 |
0.341 |
| 150 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.768 |
0.557 |
| 151 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.764 |
1.045 |
| 152 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.764 |
0.762 |
| 153 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.763 |
0.216 |
| 154 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.763 |
0.230 |
| 155 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.763 |
-0.035 |
| 156 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
0.757 |
0.140 |
| 157 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.757 |
0.058 |
| 158 |
260005_at |
AT1G67920
|
unknown |
0.756 |
0.634 |
| 159 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.755 |
0.399 |
| 160 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.753 |
0.150 |
| 161 |
265993_at |
AT2G24160
|
[pseudogene] |
0.751 |
0.094 |
| 162 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.744 |
0.563 |
| 163 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.743 |
0.421 |
| 164 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.741 |
0.359 |
| 165 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.741 |
0.394 |
| 166 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.740 |
0.341 |
| 167 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.738 |
0.008 |
| 168 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
0.737 |
0.131 |
| 169 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.737 |
0.037 |
| 170 |
264635_at |
AT1G65500
|
unknown |
0.737 |
0.095 |
| 171 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.733 |
0.076 |
| 172 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.733 |
-0.147 |
| 173 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.732 |
0.260 |
| 174 |
264951_at |
AT1G76970
|
[Target of Myb protein 1] |
0.729 |
0.584 |
| 175 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.727 |
0.491 |
| 176 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.727 |
0.340 |
| 177 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.725 |
0.674 |
| 178 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.725 |
0.155 |
| 179 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.723 |
1.033 |
| 180 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.721 |
0.015 |
| 181 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.721 |
0.500 |
| 182 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.720 |
0.658 |
| 183 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.720 |
0.126 |
| 184 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.719 |
-0.048 |
| 185 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.719 |
0.384 |
| 186 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.719 |
0.185 |
| 187 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.718 |
0.102 |
| 188 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.718 |
-0.187 |
| 189 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.717 |
0.005 |
| 190 |
266017_at |
AT2G18690
|
unknown |
0.715 |
0.383 |
| 191 |
247324_at |
AT5G64190
|
unknown |
0.715 |
-0.018 |
| 192 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.714 |
0.518 |
| 193 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.713 |
0.239 |
| 194 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.713 |
-0.023 |
| 195 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.712 |
0.112 |
| 196 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.709 |
0.433 |
| 197 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.706 |
0.214 |
| 198 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.705 |
0.292 |
| 199 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.702 |
0.287 |
| 200 |
246842_at |
AT5G26731
|
unknown |
0.701 |
0.032 |
| 201 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.700 |
0.281 |
| 202 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.699 |
0.161 |
| 203 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.699 |
0.061 |
| 204 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.698 |
0.366 |
| 205 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.697 |
-0.060 |
| 206 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.692 |
0.872 |
| 207 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.689 |
0.689 |
| 208 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.689 |
0.191 |
| 209 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.689 |
-0.007 |
| 210 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.686 |
0.283 |
| 211 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.686 |
0.046 |
| 212 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.685 |
0.188 |
| 213 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.685 |
0.445 |
| 214 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.684 |
0.510 |
| 215 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.684 |
0.052 |
| 216 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
0.682 |
0.013 |
| 217 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.678 |
0.589 |
| 218 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.677 |
0.106 |
| 219 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.676 |
0.031 |
| 220 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.675 |
1.069 |
| 221 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.673 |
0.230 |
| 222 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.670 |
0.564 |
| 223 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.670 |
0.006 |
| 224 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.669 |
0.648 |
| 225 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.663 |
0.037 |
| 226 |
245566_at |
AT4G14610
|
[pseudogene] |
0.662 |
0.436 |
| 227 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.662 |
0.387 |
| 228 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.661 |
0.010 |
| 229 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.660 |
0.694 |
| 230 |
255254_at |
AT4G05030
|
[Copper transport protein family] |
0.658 |
0.061 |
| 231 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.657 |
-0.185 |
| 232 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.657 |
0.853 |
| 233 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.653 |
0.627 |
| 234 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.651 |
0.383 |
| 235 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.651 |
0.594 |
| 236 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.650 |
0.091 |
| 237 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.649 |
-0.331 |
| 238 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.649 |
0.003 |
| 239 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.648 |
0.022 |
| 240 |
262085_at |
AT1G56060
|
unknown |
0.648 |
0.776 |
| 241 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.648 |
-0.048 |
| 242 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.647 |
0.018 |
| 243 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.647 |
0.853 |
| 244 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.646 |
-0.277 |
| 245 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.646 |
0.177 |
| 246 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.644 |
0.241 |
| 247 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.644 |
0.795 |
| 248 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.643 |
0.074 |
| 249 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.642 |
0.747 |
| 250 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.639 |
0.133 |
| 251 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.639 |
-0.109 |
| 252 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.638 |
0.185 |
| 253 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
0.638 |
-0.028 |
| 254 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.637 |
-0.064 |
| 255 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.636 |
0.373 |
| 256 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.634 |
0.173 |
| 257 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.634 |
0.041 |
| 258 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.633 |
0.375 |
| 259 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.631 |
0.213 |
| 260 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.631 |
0.881 |
| 261 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.629 |
-0.057 |
| 262 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.627 |
-0.164 |
| 263 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.623 |
-0.025 |
| 264 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.621 |
0.067 |
| 265 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.620 |
0.052 |
| 266 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.620 |
0.387 |
| 267 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.619 |
0.276 |
| 268 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.619 |
0.624 |
| 269 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.617 |
0.214 |
| 270 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.617 |
0.558 |
| 271 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.616 |
0.356 |
| 272 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.615 |
0.456 |
| 273 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.612 |
-0.083 |
| 274 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.610 |
-0.171 |
| 275 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
0.610 |
0.658 |
| 276 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.609 |
-0.180 |
| 277 |
249377_at |
AT5G40690
|
unknown |
0.609 |
0.077 |
| 278 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.608 |
0.218 |
| 279 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.607 |
0.034 |
| 280 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.606 |
0.773 |
| 281 |
249889_at |
AT5G22540
|
unknown |
0.606 |
0.590 |
| 282 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.605 |
0.434 |
| 283 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
0.605 |
0.032 |
| 284 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.603 |
0.536 |
| 285 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.602 |
-0.102 |
| 286 |
260362_at |
AT1G70530
|
CRK3, cysteine-rich RLK (RECEPTOR-like protein kinase) 3 |
0.600 |
0.340 |
| 287 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.599 |
0.051 |
| 288 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.598 |
0.076 |
| 289 |
251293_at |
AT3G61930
|
unknown |
0.597 |
0.156 |
| 290 |
249550_at |
AT5G38210
|
[Protein kinase family protein] |
0.596 |
0.239 |
| 291 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
0.595 |
0.247 |
| 292 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-1.428 |
-0.306 |
| 293 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-1.315 |
-0.235 |
| 294 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-1.110 |
-0.103 |
| 295 |
265892_at |
AT2G15020
|
unknown |
-1.040 |
-0.050 |
| 296 |
263829_at |
AT2G40435
|
unknown |
-1.039 |
-0.161 |
| 297 |
256096_at |
AT1G13650
|
unknown |
-1.012 |
-0.432 |
| 298 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.988 |
-0.257 |
| 299 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.937 |
-0.101 |
| 300 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.922 |
-0.092 |
| 301 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.919 |
-0.098 |
| 302 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.901 |
0.060 |
| 303 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.887 |
-0.308 |
| 304 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.884 |
-0.299 |
| 305 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.875 |
-0.284 |
| 306 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.864 |
0.015 |
| 307 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.863 |
0.029 |
| 308 |
254683_at |
AT4G13800
|
unknown |
-0.861 |
0.058 |
| 309 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.859 |
-0.310 |
| 310 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.852 |
-0.275 |
| 311 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.848 |
-0.068 |
| 312 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.846 |
-0.114 |
| 313 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.846 |
0.153 |
| 314 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.837 |
-0.078 |
| 315 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.815 |
-0.058 |
| 316 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.815 |
0.027 |
| 317 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.811 |
-0.207 |
| 318 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.806 |
-0.334 |
| 319 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.797 |
-0.211 |
| 320 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.796 |
-0.409 |
| 321 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.794 |
-0.091 |
| 322 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.787 |
-0.322 |
| 323 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.782 |
-0.182 |
| 324 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.782 |
-0.421 |
| 325 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.775 |
0.071 |
| 326 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.774 |
-0.313 |
| 327 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.763 |
-0.279 |
| 328 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.760 |
-0.038 |
| 329 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.759 |
-0.308 |
| 330 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.757 |
0.073 |
| 331 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.757 |
-0.151 |
| 332 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.756 |
-0.436 |
| 333 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.753 |
0.190 |
| 334 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.752 |
0.758 |
| 335 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.752 |
-0.158 |
| 336 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.745 |
-0.420 |
| 337 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.744 |
-0.140 |
| 338 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.744 |
0.046 |
| 339 |
256603_at |
AT3G28270
|
unknown |
-0.740 |
-0.157 |
| 340 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.739 |
0.025 |
| 341 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.739 |
-0.301 |
| 342 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.738 |
-0.071 |
| 343 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.737 |
0.180 |
| 344 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.737 |
-0.211 |
| 345 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.728 |
-0.196 |
| 346 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.723 |
-0.063 |
| 347 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.720 |
-0.226 |
| 348 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.718 |
0.015 |
| 349 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.716 |
-0.228 |
| 350 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.715 |
-0.214 |
| 351 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.713 |
0.103 |
| 352 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.707 |
-0.561 |
| 353 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.706 |
0.002 |
| 354 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.705 |
-0.268 |
| 355 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.705 |
-0.196 |
| 356 |
246997_at |
AT5G67390
|
unknown |
-0.699 |
-0.130 |
| 357 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.699 |
-0.189 |
| 358 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.696 |
0.077 |
| 359 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.693 |
-0.414 |
| 360 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.693 |
-0.208 |
| 361 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.691 |
-0.428 |
| 362 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.691 |
-0.418 |
| 363 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.684 |
-0.231 |
| 364 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.684 |
-0.139 |
| 365 |
254145_at |
AT4G24700
|
unknown |
-0.682 |
-0.313 |
| 366 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.681 |
0.020 |
| 367 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.680 |
-0.264 |
| 368 |
258782_at |
AT3G11750
|
FOLB1 |
-0.678 |
-0.284 |
| 369 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.677 |
-0.246 |
| 370 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.676 |
-0.132 |
| 371 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.672 |
-0.208 |
| 372 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.672 |
-0.287 |
| 373 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.670 |
-0.310 |
| 374 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
-0.670 |
-0.203 |
| 375 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.666 |
-0.046 |
| 376 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.665 |
-0.106 |
| 377 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.665 |
-0.427 |
| 378 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.664 |
-0.325 |
| 379 |
247716_at |
AT5G59350
|
unknown |
-0.658 |
-0.437 |
| 380 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.657 |
-0.190 |
| 381 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.655 |
-0.153 |
| 382 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.652 |
0.009 |
| 383 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.650 |
-0.122 |
| 384 |
251058_at |
AT5G01790
|
unknown |
-0.648 |
-0.469 |
| 385 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.646 |
-0.103 |
| 386 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.645 |
-0.144 |
| 387 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.639 |
-0.176 |
| 388 |
245743_at |
AT1G51080
|
unknown |
-0.639 |
-0.201 |
| 389 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.634 |
-0.100 |
| 390 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.633 |
0.045 |
| 391 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.632 |
-0.215 |
| 392 |
259001_at |
AT3G01960
|
unknown |
-0.631 |
0.088 |
| 393 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.627 |
0.060 |
| 394 |
253165_at |
AT4G35320
|
unknown |
-0.627 |
-0.271 |
| 395 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.626 |
-0.301 |
| 396 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.622 |
-0.248 |
| 397 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.621 |
-0.158 |
| 398 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.618 |
-0.430 |
| 399 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
-0.616 |
-0.170 |
| 400 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.616 |
-0.082 |
| 401 |
250696_at |
AT5G06790
|
unknown |
-0.615 |
-0.414 |
| 402 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-0.615 |
0.103 |
| 403 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.615 |
-0.201 |
| 404 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.614 |
-0.393 |
| 405 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.613 |
-0.406 |
| 406 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.612 |
0.043 |
| 407 |
263918_at |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
-0.610 |
-0.208 |
| 408 |
251788_at |
AT3G55420
|
unknown |
-0.610 |
-0.061 |
| 409 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.610 |
-0.365 |
| 410 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.603 |
-0.210 |
| 411 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.600 |
-0.178 |
| 412 |
247486_at |
AT5G62140
|
unknown |
-0.597 |
-0.382 |
| 413 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.596 |
-0.391 |
| 414 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.595 |
-0.155 |
| 415 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.591 |
-0.382 |
| 416 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.591 |
-0.391 |
| 417 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.590 |
-0.132 |
| 418 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.588 |
-0.185 |
| 419 |
247055_at |
AT5G66740
|
unknown |
-0.587 |
-0.100 |
| 420 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
-0.586 |
0.149 |
| 421 |
252501_at |
AT3G46880
|
unknown |
-0.585 |
0.077 |
| 422 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.584 |
0.050 |
| 423 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.583 |
-0.108 |
| 424 |
266278_at |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.582 |
-0.246 |
| 425 |
254693_at |
AT4G17880
|
MYC4 |
-0.581 |
-0.396 |
| 426 |
260112_at |
AT1G63310
|
unknown |
-0.581 |
0.114 |
| 427 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.581 |
-0.228 |
| 428 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.579 |
-0.176 |
| 429 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.579 |
0.251 |
| 430 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.578 |
-0.376 |
| 431 |
251753_at |
AT3G55760
|
unknown |
-0.578 |
-0.154 |
| 432 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.577 |
-0.193 |
| 433 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.574 |
-0.189 |
| 434 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.574 |
-0.180 |
| 435 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.568 |
-0.163 |
| 436 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.566 |
0.032 |
| 437 |
249932_at |
AT5G22390
|
unknown |
-0.565 |
-0.196 |
| 438 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.564 |
0.123 |
| 439 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.563 |
-0.152 |
| 440 |
264314_at |
AT1G70420
|
unknown |
-0.562 |
0.595 |
| 441 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.561 |
-0.185 |
| 442 |
258807_at |
AT3G04030
|
MYR2 |
-0.559 |
-0.115 |
| 443 |
263545_at |
AT2G21560
|
unknown |
-0.557 |
-0.363 |
| 444 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
-0.554 |
0.482 |
| 445 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.553 |
-0.028 |
| 446 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.548 |
-0.104 |
| 447 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.547 |
-0.291 |
| 448 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.547 |
-0.302 |
| 449 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.546 |
-0.073 |
| 450 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.545 |
-0.169 |
| 451 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.543 |
-0.105 |
| 452 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.543 |
-0.052 |
| 453 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.542 |
0.038 |
| 454 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.541 |
-0.213 |
| 455 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.541 |
-0.002 |
| 456 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.541 |
-0.098 |
| 457 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.540 |
-0.165 |
| 458 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.540 |
-0.230 |
| 459 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.539 |
-0.040 |
| 460 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.539 |
0.087 |
| 461 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.539 |
-0.107 |
| 462 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.538 |
-0.176 |
| 463 |
248393_at |
AT5G52060
|
ATBAG1, BCL-2-associated athanogene 1, BAG1, BCL-2-associated athanogene 1 |
-0.537 |
-0.014 |
| 464 |
264738_at |
AT1G62250
|
unknown |
-0.532 |
-0.105 |
| 465 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.531 |
0.025 |
| 466 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.530 |
0.121 |
| 467 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.530 |
-0.202 |
| 468 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.529 |
0.045 |
| 469 |
262811_at |
AT1G11700
|
unknown |
-0.529 |
-0.201 |
| 470 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.526 |
-0.061 |
| 471 |
256873_at |
AT3G26310
|
CYP71B35, cytochrome P450, family 71, subfamily B, polypeptide 35 |
-0.525 |
-0.261 |
| 472 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.524 |
-0.228 |
| 473 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
-0.524 |
-0.305 |
| 474 |
250936_at |
AT5G03120
|
unknown |
-0.519 |
-0.215 |
| 475 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.518 |
-0.248 |
| 476 |
247522_at |
AT5G61340
|
unknown |
-0.517 |
-0.091 |
| 477 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.516 |
-0.493 |
| 478 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.515 |
0.079 |
| 479 |
253166_at |
AT4G35290
|
ATGLR3.2, ATGLUR2, GLR3.2, GLUTAMATE RECEPTOR 3.2, GLUR2, glutamate receptor 2 |
-0.514 |
-0.083 |
| 480 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.513 |
0.043 |
| 481 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.512 |
-0.120 |
| 482 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.512 |
0.006 |
| 483 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.511 |
-0.184 |
| 484 |
250980_at |
AT5G03130
|
unknown |
-0.510 |
-0.076 |
| 485 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.510 |
-0.357 |
| 486 |
257875_at |
AT3G17120
|
unknown |
-0.509 |
-0.069 |
| 487 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.508 |
-0.005 |
| 488 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.507 |
-0.364 |
| 489 |
257672_at |
AT3G20300
|
unknown |
-0.507 |
-0.082 |
| 490 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.507 |
-0.049 |
| 491 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.506 |
0.061 |
| 492 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.505 |
-0.059 |
| 493 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.505 |
0.017 |
| 494 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.504 |
-0.125 |
| 495 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.504 |
-0.108 |
| 496 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.504 |
0.078 |
| 497 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.503 |
-0.122 |
| 498 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
-0.503 |
-0.046 |
| 499 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.502 |
0.010 |
| 500 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.501 |
-0.152 |
| 501 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.501 |
-0.024 |
| 502 |
264096_at |
AT1G78995
|
unknown |
-0.501 |
-0.366 |
| 503 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.500 |
-0.104 |
| 504 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.497 |
-0.333 |
| 505 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.497 |
0.157 |
| 506 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.497 |
-0.219 |
| 507 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.496 |
-0.104 |
| 508 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.495 |
-0.089 |
| 509 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.494 |
-0.371 |
| 510 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.494 |
-0.091 |
| 511 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.490 |
-0.079 |
| 512 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.489 |
0.005 |
| 513 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.488 |
-0.203 |
| 514 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.488 |
-0.145 |
| 515 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.486 |
-0.059 |
| 516 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.485 |
-0.002 |
| 517 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.484 |
0.017 |
| 518 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.482 |
-0.150 |
| 519 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.482 |
-0.055 |
| 520 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.482 |
0.153 |
| 521 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.480 |
-0.122 |
| 522 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.479 |
-0.351 |
| 523 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.478 |
-0.114 |
| 524 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.477 |
-0.359 |
| 525 |
246231_at |
AT4G37080
|
unknown |
-0.474 |
-0.289 |
| 526 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.474 |
-0.270 |
| 527 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
-0.472 |
0.028 |
| 528 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
-0.472 |
-0.031 |
| 529 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.471 |
-0.240 |
| 530 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.471 |
0.024 |
| 531 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.469 |
-0.150 |
| 532 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.469 |
0.165 |
| 533 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.468 |
-0.044 |
| 534 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
-0.468 |
-0.107 |
| 535 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.467 |
-0.114 |
| 536 |
248646_at |
AT5G49100
|
unknown |
-0.467 |
-0.215 |
| 537 |
261776_at |
AT1G76190
|
SAUR56, SMALL AUXIN UPREGULATED RNA 56 |
-0.464 |
-0.015 |
| 538 |
266227_at |
AT2G28870
|
unknown |
-0.463 |
0.003 |
| 539 |
259188_at |
AT3G01510
|
LSF1, like SEX4 1 |
-0.462 |
-0.017 |
| 540 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.462 |
0.092 |
| 541 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.461 |
-0.218 |
| 542 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.461 |
-0.109 |
| 543 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.461 |
0.059 |
| 544 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.459 |
-0.138 |
| 545 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.459 |
-0.175 |
| 546 |
245601_at |
AT4G14240
|
unknown |
-0.459 |
-0.139 |
| 547 |
261921_at |
AT1G65900
|
unknown |
-0.458 |
-0.050 |
| 548 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
-0.458 |
0.081 |
| 549 |
251110_at |
AT5G01260
|
[Carbohydrate-binding-like fold] |
-0.458 |
-0.192 |
| 550 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.455 |
-0.378 |
| 551 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.455 |
0.073 |
| 552 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.455 |
-0.168 |
| 553 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.454 |
-0.108 |
| 554 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
-0.454 |
0.098 |
| 555 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.453 |
-0.403 |
| 556 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.452 |
0.051 |
| 557 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.452 |
-0.269 |
| 558 |
256529_at |
AT1G33260
|
[Protein kinase superfamily protein] |
-0.451 |
0.031 |
| 559 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.451 |
0.093 |
| 560 |
249303_at |
AT5G41460
|
unknown |
-0.451 |
-0.065 |
| 561 |
257275_at |
AT3G14450
|
CID9, CTC-interacting domain 9 |
-0.449 |
0.095 |
| 562 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.449 |
-0.107 |
| 563 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.449 |
0.073 |
| 564 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.449 |
-0.394 |
| 565 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.449 |
0.214 |
| 566 |
255854_at |
AT1G67050
|
unknown |
-0.448 |
0.286 |
| 567 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.448 |
-0.147 |
| 568 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.448 |
-0.168 |
| 569 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.447 |
-0.004 |
| 570 |
251219_at |
AT3G62390
|
TBL6, TRICHOME BIREFRINGENCE-LIKE 6 |
-0.445 |
-0.155 |
| 571 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.444 |
-0.212 |
| 572 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.444 |
-0.084 |
| 573 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
-0.444 |
-0.303 |
| 574 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.443 |
-0.070 |
| 575 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.442 |
-0.259 |
| 576 |
265327_at |
AT2G18210
|
unknown |
-0.442 |
0.334 |
| 577 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
-0.441 |
-0.054 |
| 578 |
260696_at |
AT1G32520
|
unknown |
-0.440 |
-0.228 |
| 579 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
-0.439 |
-0.045 |
| 580 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.438 |
-0.381 |
| 581 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.437 |
-0.048 |