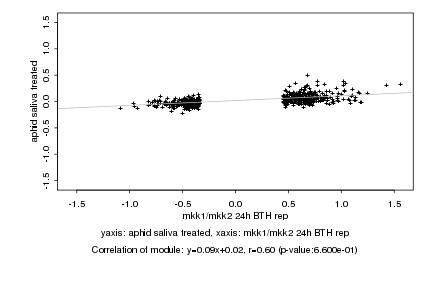
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
mkk1/mkk2 24h BTH rep |
Log2 signal ratio
aphid saliva treated |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.556 |
0.331 |
| 2 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.421 |
0.321 |
| 3 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.243 |
0.154 |
| 4 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.189 |
-0.012 |
| 5 |
267345_at |
AT2G44240
|
unknown |
1.177 |
-0.013 |
| 6 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.166 |
0.153 |
| 7 |
245076_at |
AT2G23170
|
GH3.3 |
1.155 |
0.178 |
| 8 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.134 |
0.078 |
| 9 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
1.128 |
0.022 |
| 10 |
251635_at |
AT3G57510
|
ADPG1 |
1.119 |
0.021 |
| 11 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
1.104 |
0.238 |
| 12 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.100 |
0.121 |
| 13 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.089 |
0.097 |
| 14 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
1.078 |
-0.033 |
| 15 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
1.075 |
0.037 |
| 16 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
1.068 |
0.039 |
| 17 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
1.039 |
0.345 |
| 18 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
1.028 |
0.223 |
| 19 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.024 |
0.197 |
| 20 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
1.019 |
0.396 |
| 21 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
1.017 |
0.053 |
| 22 |
249454_at |
AT5G39520
|
unknown |
1.014 |
0.319 |
| 23 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.999 |
0.146 |
| 24 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.972 |
0.008 |
| 25 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.968 |
0.156 |
| 26 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.960 |
0.035 |
| 27 |
264580_at |
AT1G05340
|
unknown |
0.958 |
0.062 |
| 28 |
260668_at |
AT1G19530
|
unknown |
0.958 |
0.100 |
| 29 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.946 |
0.259 |
| 30 |
251612_at |
AT3G57950
|
unknown |
0.924 |
0.019 |
| 31 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.921 |
-0.030 |
| 32 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.916 |
0.054 |
| 33 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.914 |
0.159 |
| 34 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.908 |
-0.011 |
| 35 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.896 |
0.094 |
| 36 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.888 |
-0.055 |
| 37 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.881 |
0.196 |
| 38 |
263475_at |
AT2G31945
|
unknown |
0.866 |
0.030 |
| 39 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.865 |
0.034 |
| 40 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.861 |
0.071 |
| 41 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.856 |
0.209 |
| 42 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.855 |
0.128 |
| 43 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.852 |
-0.023 |
| 44 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
0.848 |
0.089 |
| 45 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.841 |
0.123 |
| 46 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.839 |
0.339 |
| 47 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.834 |
0.060 |
| 48 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.826 |
0.066 |
| 49 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.825 |
0.025 |
| 50 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.825 |
0.084 |
| 51 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.824 |
0.055 |
| 52 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.820 |
0.029 |
| 53 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.818 |
0.169 |
| 54 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.815 |
0.042 |
| 55 |
267178_at |
AT2G37750
|
unknown |
0.807 |
0.066 |
| 56 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.802 |
0.114 |
| 57 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.800 |
0.028 |
| 58 |
251293_at |
AT3G61930
|
unknown |
0.800 |
0.012 |
| 59 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.785 |
0.196 |
| 60 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.779 |
0.005 |
| 61 |
264680_at |
AT1G65510
|
unknown |
0.778 |
0.004 |
| 62 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.775 |
0.316 |
| 63 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.772 |
0.383 |
| 64 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.770 |
0.064 |
| 65 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.769 |
0.117 |
| 66 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.767 |
0.077 |
| 67 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
0.762 |
0.015 |
| 68 |
253827_at |
AT4G28085
|
unknown |
0.760 |
0.124 |
| 69 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.759 |
0.041 |
| 70 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.756 |
0.187 |
| 71 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.754 |
0.066 |
| 72 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.749 |
0.136 |
| 73 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.747 |
0.048 |
| 74 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.745 |
0.067 |
| 75 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.744 |
0.004 |
| 76 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.739 |
0.179 |
| 77 |
249004_at |
AT5G44570
|
unknown |
0.736 |
0.079 |
| 78 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.734 |
-0.009 |
| 79 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.731 |
0.156 |
| 80 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.731 |
0.036 |
| 81 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.724 |
0.033 |
| 82 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.722 |
0.001 |
| 83 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.720 |
0.123 |
| 84 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.720 |
-0.064 |
| 85 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.718 |
-0.051 |
| 86 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.717 |
-0.007 |
| 87 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
0.716 |
0.080 |
| 88 |
255912_at |
AT1G66960
|
LUP5 |
0.716 |
0.047 |
| 89 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.715 |
0.036 |
| 90 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.713 |
0.100 |
| 91 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.712 |
0.157 |
| 92 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.712 |
-0.052 |
| 93 |
260522_x_at |
AT2G41730
|
unknown |
0.711 |
0.050 |
| 94 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.706 |
0.176 |
| 95 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.704 |
0.192 |
| 96 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.704 |
0.191 |
| 97 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.703 |
0.171 |
| 98 |
261243_at |
AT1G20180
|
unknown |
0.702 |
0.049 |
| 99 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.699 |
0.047 |
| 100 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
0.697 |
0.004 |
| 101 |
258209_at |
AT3G14060
|
unknown |
0.697 |
0.165 |
| 102 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.696 |
-0.057 |
| 103 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.693 |
0.108 |
| 104 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.693 |
-0.025 |
| 105 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.692 |
0.248 |
| 106 |
259596_at |
AT1G28130
|
GH3.17 |
0.692 |
0.040 |
| 107 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.691 |
0.103 |
| 108 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.689 |
0.030 |
| 109 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.688 |
0.142 |
| 110 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.687 |
-0.041 |
| 111 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.684 |
0.063 |
| 112 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.682 |
0.136 |
| 113 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.679 |
0.309 |
| 114 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.677 |
0.072 |
| 115 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.677 |
0.027 |
| 116 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.676 |
0.028 |
| 117 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.676 |
0.495 |
| 118 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.675 |
-0.002 |
| 119 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.672 |
0.162 |
| 120 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
0.671 |
0.083 |
| 121 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.671 |
0.073 |
| 122 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.670 |
0.052 |
| 123 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.670 |
0.287 |
| 124 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
0.670 |
0.003 |
| 125 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.666 |
-0.004 |
| 126 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
0.665 |
-0.003 |
| 127 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
0.662 |
0.038 |
| 128 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.662 |
0.269 |
| 129 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.661 |
-0.046 |
| 130 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.659 |
0.131 |
| 131 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.658 |
0.061 |
| 132 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.657 |
0.152 |
| 133 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.652 |
0.134 |
| 134 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.650 |
-0.029 |
| 135 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.650 |
0.215 |
| 136 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.650 |
0.236 |
| 137 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.649 |
-0.004 |
| 138 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.648 |
0.056 |
| 139 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.648 |
0.049 |
| 140 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.648 |
0.277 |
| 141 |
267226_at |
AT2G44010
|
unknown |
0.647 |
-0.013 |
| 142 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.647 |
-0.017 |
| 143 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.645 |
0.051 |
| 144 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.644 |
0.141 |
| 145 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.642 |
0.090 |
| 146 |
249896_at |
AT5G22530
|
unknown |
0.640 |
0.036 |
| 147 |
247913_at |
AT5G57510
|
unknown |
0.639 |
-0.097 |
| 148 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.636 |
0.176 |
| 149 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.633 |
0.113 |
| 150 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.626 |
0.143 |
| 151 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.625 |
0.073 |
| 152 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.623 |
-0.053 |
| 153 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.622 |
0.076 |
| 154 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.621 |
0.057 |
| 155 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.620 |
0.232 |
| 156 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
0.618 |
-0.000 |
| 157 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.616 |
-0.018 |
| 158 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.616 |
0.060 |
| 159 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
0.613 |
-0.001 |
| 160 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.611 |
-0.008 |
| 161 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.610 |
0.089 |
| 162 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.609 |
0.043 |
| 163 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.608 |
0.117 |
| 164 |
258203_at |
AT3G13950
|
unknown |
0.607 |
0.016 |
| 165 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.607 |
0.034 |
| 166 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.607 |
0.185 |
| 167 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.607 |
0.123 |
| 168 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.606 |
0.098 |
| 169 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.606 |
0.103 |
| 170 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
0.603 |
0.041 |
| 171 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.601 |
0.087 |
| 172 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.600 |
0.199 |
| 173 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.600 |
0.053 |
| 174 |
247324_at |
AT5G64190
|
unknown |
0.599 |
0.116 |
| 175 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.597 |
-0.042 |
| 176 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.596 |
0.082 |
| 177 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.593 |
-0.001 |
| 178 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.592 |
-0.030 |
| 179 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.591 |
0.167 |
| 180 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.590 |
-0.039 |
| 181 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.589 |
0.020 |
| 182 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.589 |
0.119 |
| 183 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.588 |
-0.002 |
| 184 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.587 |
0.122 |
| 185 |
251419_at |
AT3G60470
|
unknown |
0.586 |
-0.024 |
| 186 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.584 |
-0.019 |
| 187 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.582 |
0.142 |
| 188 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.582 |
0.040 |
| 189 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.580 |
0.117 |
| 190 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.577 |
0.066 |
| 191 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.574 |
0.107 |
| 192 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.572 |
0.088 |
| 193 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.571 |
0.090 |
| 194 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
0.571 |
0.033 |
| 195 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.570 |
0.023 |
| 196 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.570 |
0.061 |
| 197 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.570 |
0.167 |
| 198 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.569 |
0.144 |
| 199 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.569 |
0.087 |
| 200 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.569 |
0.108 |
| 201 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.568 |
0.079 |
| 202 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.567 |
0.067 |
| 203 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
0.565 |
0.042 |
| 204 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.565 |
0.034 |
| 205 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.563 |
0.051 |
| 206 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.562 |
0.353 |
| 207 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.561 |
0.027 |
| 208 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.559 |
0.068 |
| 209 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.557 |
0.167 |
| 210 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.555 |
0.055 |
| 211 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.555 |
0.073 |
| 212 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.554 |
0.108 |
| 213 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.554 |
0.040 |
| 214 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.551 |
-0.010 |
| 215 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.550 |
0.057 |
| 216 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.548 |
0.155 |
| 217 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.547 |
0.102 |
| 218 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.543 |
-0.065 |
| 219 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.543 |
0.128 |
| 220 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.543 |
0.189 |
| 221 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.542 |
0.116 |
| 222 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.541 |
-0.022 |
| 223 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.541 |
-0.037 |
| 224 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.533 |
0.092 |
| 225 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.533 |
-0.004 |
| 226 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.527 |
0.127 |
| 227 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.527 |
0.074 |
| 228 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
0.525 |
0.147 |
| 229 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
0.522 |
0.117 |
| 230 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.521 |
0.090 |
| 231 |
260744_at |
AT1G15010
|
unknown |
0.521 |
-0.015 |
| 232 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.517 |
-0.006 |
| 233 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.516 |
0.145 |
| 234 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.516 |
0.002 |
| 235 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.516 |
0.027 |
| 236 |
246406_at |
AT1G57650
|
[ATP binding] |
0.511 |
0.030 |
| 237 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.510 |
0.058 |
| 238 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.510 |
0.064 |
| 239 |
256789_at |
AT3G13672
|
SINA2 |
0.508 |
0.287 |
| 240 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.507 |
0.028 |
| 241 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.505 |
0.079 |
| 242 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.504 |
0.176 |
| 243 |
251766_at |
AT3G55910
|
unknown |
0.502 |
0.003 |
| 244 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.500 |
0.023 |
| 245 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.497 |
0.005 |
| 246 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.496 |
0.070 |
| 247 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.496 |
0.065 |
| 248 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.495 |
0.086 |
| 249 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.490 |
0.022 |
| 250 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.490 |
-0.013 |
| 251 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.489 |
0.011 |
| 252 |
254598_at |
AT4G18990
|
XTH29, xyloglucan endotransglucosylase/hydrolase 29 |
0.488 |
-0.012 |
| 253 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.488 |
0.025 |
| 254 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.487 |
0.060 |
| 255 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.486 |
0.070 |
| 256 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.486 |
-0.053 |
| 257 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.485 |
0.068 |
| 258 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.485 |
0.145 |
| 259 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.484 |
0.081 |
| 260 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.483 |
0.001 |
| 261 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.482 |
0.194 |
| 262 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.482 |
-0.065 |
| 263 |
260243_at |
AT1G63720
|
unknown |
0.482 |
-0.021 |
| 264 |
251019_at |
AT5G02420
|
unknown |
0.482 |
0.057 |
| 265 |
256803_at |
AT3G20960
|
CYP705A33, cytochrome P450, family 705, subfamily A, polypeptide 33 |
0.481 |
0.000 |
| 266 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.480 |
0.105 |
| 267 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.479 |
0.030 |
| 268 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.478 |
0.030 |
| 269 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.478 |
-0.029 |
| 270 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.477 |
0.193 |
| 271 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.477 |
0.059 |
| 272 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
0.477 |
0.057 |
| 273 |
255430_at |
AT4G03320
|
AtTic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV, Tic20-IV, translocon at the inner envelope membrane of chloroplasts 20-IV |
0.476 |
0.066 |
| 274 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.476 |
-0.001 |
| 275 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.475 |
0.060 |
| 276 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.473 |
0.079 |
| 277 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.473 |
0.003 |
| 278 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.472 |
0.027 |
| 279 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.472 |
0.228 |
| 280 |
267624_at |
AT2G39660
|
BIK1, botrytis-induced kinase1 |
0.471 |
0.050 |
| 281 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.469 |
-0.095 |
| 282 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.468 |
0.001 |
| 283 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.468 |
0.055 |
| 284 |
249897_at |
AT5G22550
|
unknown |
0.466 |
0.003 |
| 285 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.465 |
-0.074 |
| 286 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.465 |
-0.049 |
| 287 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.462 |
0.089 |
| 288 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.462 |
0.105 |
| 289 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.461 |
-0.010 |
| 290 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.460 |
0.112 |
| 291 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.460 |
0.051 |
| 292 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.459 |
0.096 |
| 293 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
0.458 |
-0.031 |
| 294 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.457 |
0.064 |
| 295 |
246909_at |
AT5G25770
|
[alpha/beta-Hydrolases superfamily protein] |
0.454 |
0.070 |
| 296 |
266571_at |
AT2G23830
|
[PapD-like superfamily protein] |
0.453 |
-0.009 |
| 297 |
254736_at |
AT4G13820
|
[Leucine-rich repeat (LRR) family protein] |
0.453 |
0.009 |
| 298 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.453 |
0.106 |
| 299 |
256930_at |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
0.453 |
0.131 |
| 300 |
263118_at |
AT1G03090
|
MCCA |
0.452 |
0.048 |
| 301 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-1.088 |
-0.131 |
| 302 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.969 |
-0.019 |
| 303 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.955 |
-0.079 |
| 304 |
256096_at |
AT1G13650
|
unknown |
-0.928 |
-0.122 |
| 305 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.829 |
-0.058 |
| 306 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.824 |
0.019 |
| 307 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.811 |
-0.051 |
| 308 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.792 |
-0.001 |
| 309 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.771 |
0.020 |
| 310 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.767 |
-0.092 |
| 311 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.765 |
-0.071 |
| 312 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.764 |
0.010 |
| 313 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.747 |
-0.109 |
| 314 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.744 |
0.002 |
| 315 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.740 |
-0.066 |
| 316 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.733 |
-0.029 |
| 317 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.726 |
0.022 |
| 318 |
263829_at |
AT2G40435
|
unknown |
-0.715 |
0.107 |
| 319 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.713 |
0.004 |
| 320 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.709 |
-0.020 |
| 321 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.693 |
-0.106 |
| 322 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.674 |
-0.029 |
| 323 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.673 |
-0.046 |
| 324 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.666 |
0.005 |
| 325 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.666 |
-0.016 |
| 326 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.657 |
-0.058 |
| 327 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.657 |
-0.013 |
| 328 |
259001_at |
AT3G01960
|
unknown |
-0.656 |
-0.016 |
| 329 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.650 |
-0.041 |
| 330 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.646 |
0.058 |
| 331 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.632 |
-0.046 |
| 332 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.632 |
-0.033 |
| 333 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.626 |
-0.030 |
| 334 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
-0.626 |
-0.040 |
| 335 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.625 |
-0.019 |
| 336 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.624 |
-0.102 |
| 337 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.610 |
-0.033 |
| 338 |
249932_at |
AT5G22390
|
unknown |
-0.609 |
-0.043 |
| 339 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.607 |
-0.171 |
| 340 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.606 |
-0.044 |
| 341 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.605 |
-0.025 |
| 342 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.604 |
-0.051 |
| 343 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.603 |
-0.093 |
| 344 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.599 |
-0.006 |
| 345 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.596 |
-0.023 |
| 346 |
254145_at |
AT4G24700
|
unknown |
-0.595 |
-0.057 |
| 347 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.593 |
0.008 |
| 348 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.591 |
-0.121 |
| 349 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
-0.586 |
-0.073 |
| 350 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.586 |
-0.002 |
| 351 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.585 |
-0.083 |
| 352 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.585 |
-0.005 |
| 353 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.584 |
-0.056 |
| 354 |
251058_at |
AT5G01790
|
unknown |
-0.584 |
-0.120 |
| 355 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.583 |
0.034 |
| 356 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.582 |
-0.077 |
| 357 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.578 |
-0.116 |
| 358 |
252501_at |
AT3G46880
|
unknown |
-0.576 |
-0.024 |
| 359 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.572 |
0.072 |
| 360 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.560 |
-0.089 |
| 361 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.556 |
-0.087 |
| 362 |
247486_at |
AT5G62140
|
unknown |
-0.555 |
-0.027 |
| 363 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.553 |
-0.002 |
| 364 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.550 |
-0.037 |
| 365 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.550 |
-0.047 |
| 366 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.549 |
-0.002 |
| 367 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.545 |
-0.066 |
| 368 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.544 |
-0.028 |
| 369 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.543 |
-0.005 |
| 370 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.541 |
-0.013 |
| 371 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.538 |
-0.038 |
| 372 |
251788_at |
AT3G55420
|
unknown |
-0.535 |
-0.050 |
| 373 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.532 |
0.052 |
| 374 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.532 |
-0.057 |
| 375 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.529 |
-0.037 |
| 376 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.518 |
-0.058 |
| 377 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.514 |
0.038 |
| 378 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.514 |
-0.003 |
| 379 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.513 |
-0.054 |
| 380 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.512 |
-0.027 |
| 381 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.511 |
-0.067 |
| 382 |
260112_at |
AT1G63310
|
unknown |
-0.507 |
-0.217 |
| 383 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.503 |
-0.041 |
| 384 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.501 |
0.019 |
| 385 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.500 |
-0.055 |
| 386 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
-0.500 |
-0.010 |
| 387 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.496 |
0.041 |
| 388 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.494 |
-0.057 |
| 389 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.491 |
0.016 |
| 390 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.489 |
0.026 |
| 391 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.488 |
-0.100 |
| 392 |
256872_at |
AT3G26490
|
[Phototropic-responsive NPH3 family protein] |
-0.485 |
0.059 |
| 393 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.485 |
-0.066 |
| 394 |
249726_at |
AT5G35480
|
unknown |
-0.485 |
0.065 |
| 395 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.484 |
-0.040 |
| 396 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
-0.484 |
-0.134 |
| 397 |
262162_at |
AT1G78020
|
unknown |
-0.483 |
0.016 |
| 398 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.482 |
-0.040 |
| 399 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
-0.482 |
-0.009 |
| 400 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.481 |
0.016 |
| 401 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.478 |
-0.003 |
| 402 |
255028_at |
AT4G09890
|
unknown |
-0.478 |
0.002 |
| 403 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.477 |
-0.020 |
| 404 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.477 |
-0.109 |
| 405 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.475 |
-0.078 |
| 406 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.474 |
0.041 |
| 407 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.474 |
-0.045 |
| 408 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
-0.473 |
0.050 |
| 409 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.469 |
0.008 |
| 410 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.469 |
-0.045 |
| 411 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.468 |
-0.036 |
| 412 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.468 |
0.006 |
| 413 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.467 |
-0.044 |
| 414 |
258468_at |
AT3G06070
|
unknown |
-0.466 |
0.025 |
| 415 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.466 |
0.040 |
| 416 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
-0.466 |
-0.002 |
| 417 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.466 |
0.014 |
| 418 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.465 |
-0.135 |
| 419 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.463 |
-0.005 |
| 420 |
264738_at |
AT1G62250
|
unknown |
-0.462 |
-0.001 |
| 421 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.461 |
-0.057 |
| 422 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.461 |
0.034 |
| 423 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.460 |
-0.112 |
| 424 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.459 |
-0.083 |
| 425 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.459 |
-0.057 |
| 426 |
247716_at |
AT5G59350
|
unknown |
-0.458 |
-0.062 |
| 427 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.458 |
-0.006 |
| 428 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.458 |
-0.021 |
| 429 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.457 |
0.023 |
| 430 |
254683_at |
AT4G13800
|
unknown |
-0.456 |
0.000 |
| 431 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.454 |
0.017 |
| 432 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.454 |
-0.033 |
| 433 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.452 |
-0.004 |
| 434 |
247331_at |
AT5G63530
|
ATFP3, ARABIDOPSIS THALIANA FARNESYLATED PROTEIN 3, FP3, farnesylated protein 3 |
-0.452 |
0.012 |
| 435 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.451 |
-0.017 |
| 436 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.451 |
-0.024 |
| 437 |
258782_at |
AT3G11750
|
FOLB1 |
-0.451 |
-0.013 |
| 438 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
-0.449 |
-0.013 |
| 439 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
-0.448 |
0.018 |
| 440 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.447 |
-0.067 |
| 441 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.447 |
-0.036 |
| 442 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.447 |
0.096 |
| 443 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.446 |
-0.022 |
| 444 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.445 |
-0.042 |
| 445 |
250696_at |
AT5G06790
|
unknown |
-0.445 |
-0.116 |
| 446 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.444 |
-0.001 |
| 447 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.443 |
-0.082 |
| 448 |
266227_at |
AT2G28870
|
unknown |
-0.442 |
-0.084 |
| 449 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.440 |
0.002 |
| 450 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.439 |
0.016 |
| 451 |
265892_at |
AT2G15020
|
unknown |
-0.439 |
-0.111 |
| 452 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.439 |
0.027 |
| 453 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
-0.438 |
0.037 |
| 454 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.437 |
-0.154 |
| 455 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.436 |
0.079 |
| 456 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.436 |
-0.002 |
| 457 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.436 |
-0.015 |
| 458 |
259765_at |
AT1G64370
|
unknown |
-0.435 |
0.006 |
| 459 |
264693_at |
AT1G69970
|
CLE26, CLAVATA3/ESR-RELATED 26 |
-0.435 |
-0.075 |
| 460 |
253243_at |
AT4G34560
|
unknown |
-0.434 |
-0.018 |
| 461 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.434 |
-0.016 |
| 462 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.434 |
0.004 |
| 463 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
-0.433 |
-0.031 |
| 464 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.433 |
-0.060 |
| 465 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.433 |
-0.074 |
| 466 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.432 |
0.002 |
| 467 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.432 |
-0.058 |
| 468 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.432 |
0.063 |
| 469 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.432 |
0.019 |
| 470 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.431 |
0.023 |
| 471 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.431 |
-0.039 |
| 472 |
250881_at |
AT5G04080
|
unknown |
-0.431 |
-0.079 |
| 473 |
250936_at |
AT5G03120
|
unknown |
-0.430 |
-0.026 |
| 474 |
245743_at |
AT1G51080
|
unknown |
-0.428 |
-0.060 |
| 475 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.428 |
-0.065 |
| 476 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.428 |
-0.037 |
| 477 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.428 |
-0.032 |
| 478 |
263352_at |
AT2G22080
|
unknown |
-0.427 |
0.050 |
| 479 |
267538_at |
AT2G41870
|
[Remorin family protein] |
-0.423 |
0.051 |
| 480 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.422 |
-0.065 |
| 481 |
267367_at |
AT2G44210
|
unknown |
-0.421 |
-0.021 |
| 482 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.421 |
-0.045 |
| 483 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.419 |
-0.010 |
| 484 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.418 |
-0.014 |
| 485 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.417 |
0.062 |
| 486 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.415 |
0.005 |
| 487 |
256569_at |
AT3G19550
|
unknown |
-0.414 |
-0.005 |
| 488 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
-0.414 |
-0.083 |
| 489 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
-0.413 |
-0.069 |
| 490 |
253165_at |
AT4G35320
|
unknown |
-0.412 |
0.034 |
| 491 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.412 |
-0.023 |
| 492 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.411 |
-0.118 |
| 493 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.410 |
-0.016 |
| 494 |
255957_at |
AT1G22160
|
unknown |
-0.410 |
0.076 |
| 495 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
-0.408 |
-0.096 |
| 496 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.406 |
-0.034 |
| 497 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.405 |
-0.023 |
| 498 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.404 |
0.018 |
| 499 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.403 |
-0.065 |
| 500 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.402 |
-0.004 |
| 501 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.401 |
-0.002 |
| 502 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.401 |
-0.041 |
| 503 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.400 |
0.003 |
| 504 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.400 |
-0.044 |
| 505 |
256293_at |
AT1G69440
|
AGO7, ARGONAUTE7, ZIP, ZIPPY |
-0.400 |
-0.028 |
| 506 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.399 |
0.043 |
| 507 |
252025_at |
AT3G52900
|
unknown |
-0.399 |
-0.102 |
| 508 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.399 |
0.003 |
| 509 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.398 |
-0.026 |
| 510 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.398 |
0.015 |
| 511 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.398 |
-0.019 |
| 512 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.397 |
0.128 |
| 513 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
-0.397 |
-0.037 |
| 514 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.396 |
-0.021 |
| 515 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.395 |
-0.005 |
| 516 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.392 |
-0.058 |
| 517 |
250732_at |
AT5G06480
|
[Immunoglobulin E-set superfamily protein] |
-0.392 |
-0.037 |
| 518 |
247754_at |
AT5G59080
|
unknown |
-0.391 |
0.028 |
| 519 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
-0.391 |
-0.009 |
| 520 |
248923_at |
AT5G45940
|
AtNUDT11, Arabidopsis thaliana nudix hydrolase homolog 11, AtNUDX11, Arabidopsis thaliana nudix hydrolase homolog 11, NUDT11, nudix hydrolase homolog 11, NUDX11, nudix hydrolase homolog 11 |
-0.391 |
-0.008 |
| 521 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.390 |
-0.074 |
| 522 |
246997_at |
AT5G67390
|
unknown |
-0.390 |
-0.106 |
| 523 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
-0.389 |
-0.025 |
| 524 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.389 |
-0.002 |
| 525 |
252073_at |
AT3G51750
|
unknown |
-0.389 |
-0.115 |
| 526 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.387 |
-0.081 |
| 527 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.387 |
0.044 |
| 528 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.385 |
-0.035 |
| 529 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.383 |
-0.094 |
| 530 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.383 |
0.009 |
| 531 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.382 |
-0.017 |
| 532 |
251219_at |
AT3G62390
|
TBL6, TRICHOME BIREFRINGENCE-LIKE 6 |
-0.382 |
-0.027 |
| 533 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.382 |
-0.011 |
| 534 |
252215_at |
AT3G50240
|
KICP-02 |
-0.380 |
-0.048 |
| 535 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.380 |
-0.040 |
| 536 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.379 |
-0.067 |
| 537 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.377 |
-0.018 |
| 538 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.376 |
-0.023 |
| 539 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.376 |
-0.033 |
| 540 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.375 |
-0.013 |
| 541 |
253294_at |
AT4G33740
|
unknown |
-0.375 |
0.040 |
| 542 |
262438_at |
AT1G47410
|
unknown |
-0.375 |
-0.107 |
| 543 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.375 |
-0.066 |
| 544 |
249167_at |
AT5G42860
|
unknown |
-0.375 |
0.003 |
| 545 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.374 |
0.003 |
| 546 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.374 |
0.017 |
| 547 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.374 |
0.019 |
| 548 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.373 |
-0.071 |
| 549 |
257761_at |
AT3G23090
|
WDL3 |
-0.373 |
0.025 |
| 550 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.373 |
-0.023 |
| 551 |
260696_at |
AT1G32520
|
unknown |
-0.371 |
-0.009 |
| 552 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.368 |
-0.012 |
| 553 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.367 |
0.000 |
| 554 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.367 |
-0.017 |
| 555 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.367 |
0.032 |
| 556 |
248172_at |
AT5G54660
|
[HSP20-like chaperones superfamily protein] |
-0.366 |
-0.060 |
| 557 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.366 |
0.025 |
| 558 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.366 |
0.006 |
| 559 |
249303_at |
AT5G41460
|
unknown |
-0.364 |
0.026 |
| 560 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.364 |
-0.001 |
| 561 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.363 |
-0.041 |
| 562 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.363 |
-0.090 |
| 563 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.362 |
0.002 |
| 564 |
255824_at |
AT2G40530
|
unknown |
-0.361 |
0.001 |
| 565 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.361 |
-0.026 |
| 566 |
263597_at |
AT2G01870
|
unknown |
-0.361 |
-0.042 |
| 567 |
245254_at |
AT4G14680
|
APS3 |
-0.361 |
-0.000 |
| 568 |
249332_at |
AT5G40980
|
unknown |
-0.360 |
-0.100 |
| 569 |
246487_at |
AT5G16030
|
unknown |
-0.358 |
0.032 |
| 570 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.358 |
0.009 |
| 571 |
249811_at |
AT5G23760
|
[Copper transport protein family] |
-0.358 |
0.018 |
| 572 |
255854_at |
AT1G67050
|
unknown |
-0.357 |
0.011 |
| 573 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.357 |
-0.050 |
| 574 |
264314_at |
AT1G70420
|
unknown |
-0.356 |
-0.068 |
| 575 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.356 |
-0.022 |
| 576 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.356 |
-0.029 |
| 577 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
-0.355 |
0.140 |
| 578 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
-0.355 |
-0.048 |
| 579 |
249134_at |
AT5G43150
|
unknown |
-0.354 |
-0.010 |
| 580 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.354 |
0.014 |
| 581 |
266273_at |
AT2G29410
|
ATMTPB1, MTPB1, metal tolerance protein B1 |
-0.354 |
0.010 |
| 582 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.354 |
-0.059 |
| 583 |
257502_at |
AT1G78110
|
unknown |
-0.353 |
0.089 |
| 584 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.353 |
-0.083 |
| 585 |
260012_at |
AT1G67865
|
unknown |
-0.352 |
0.004 |
| 586 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.352 |
-0.001 |
| 587 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.352 |
0.060 |
| 588 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
-0.352 |
-0.007 |
| 589 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.351 |
-0.041 |
| 590 |
246792_at |
AT5G27290
|
unknown |
-0.350 |
-0.031 |
| 591 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.350 |
-0.134 |
| 592 |
247522_at |
AT5G61340
|
unknown |
-0.349 |
-0.028 |
| 593 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
-0.349 |
-0.022 |
| 594 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.349 |
0.077 |
| 595 |
267060_at |
AT2G32580
|
unknown |
-0.349 |
-0.019 |
| 596 |
263674_at |
AT2G04790
|
unknown |
-0.348 |
-0.021 |
| 597 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.348 |
0.035 |
| 598 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
-0.348 |
0.045 |
| 599 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
-0.347 |
-0.000 |
| 600 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-0.347 |
0.026 |