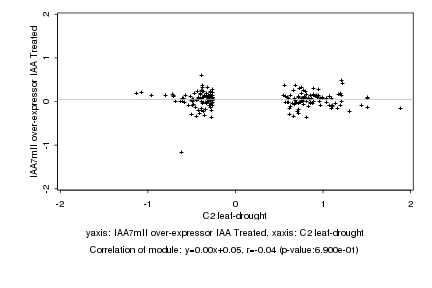
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
C2 leaf-drought |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.880 |
-0.141 |
| 2 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.507 |
-0.132 |
| 3 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.504 |
0.115 |
| 4 |
264580_at |
AT1G05340
|
unknown |
1.504 |
0.081 |
| 5 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.433 |
-0.071 |
| 6 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
1.292 |
-0.211 |
| 7 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.221 |
0.429 |
| 8 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
1.205 |
0.023 |
| 9 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
1.203 |
0.483 |
| 10 |
249039_at |
AT5G44310
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.201 |
0.152 |
| 11 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
1.191 |
-0.088 |
| 12 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.190 |
0.202 |
| 13 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.176 |
0.161 |
| 14 |
247061_at |
AT5G66780
|
unknown |
1.162 |
-0.159 |
| 15 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.138 |
-0.036 |
| 16 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
1.099 |
-0.104 |
| 17 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
1.097 |
-0.079 |
| 18 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
1.092 |
-0.156 |
| 19 |
263881_at |
AT2G21820
|
unknown |
1.091 |
-0.111 |
| 20 |
263544_at |
AT2G21590
|
APL4 |
1.086 |
0.079 |
| 21 |
253595_at |
AT4G30830
|
unknown |
1.070 |
0.134 |
| 22 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
1.064 |
-0.084 |
| 23 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
1.063 |
0.128 |
| 24 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
1.038 |
0.090 |
| 25 |
253494_at |
AT4G31830
|
unknown |
1.034 |
-0.010 |
| 26 |
256789_at |
AT3G13672
|
SINA2 |
0.984 |
0.079 |
| 27 |
254823_at |
AT4G12580
|
unknown |
0.983 |
0.094 |
| 28 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.971 |
0.075 |
| 29 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.962 |
-0.091 |
| 30 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.953 |
0.012 |
| 31 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.941 |
0.294 |
| 32 |
265024_at |
AT1G24600
|
unknown |
0.941 |
0.159 |
| 33 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.935 |
0.120 |
| 34 |
245160_at |
AT2G33080
|
AtRLP28, receptor like protein 28, RLP28, receptor like protein 28 |
0.927 |
0.094 |
| 35 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.915 |
0.144 |
| 36 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.905 |
0.119 |
| 37 |
248218_at |
AT5G53710
|
unknown |
0.890 |
0.141 |
| 38 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.888 |
0.006 |
| 39 |
259217_at |
AT3G03620
|
[MATE efflux family protein] |
0.887 |
0.168 |
| 40 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.887 |
0.299 |
| 41 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.876 |
-0.037 |
| 42 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
0.859 |
0.072 |
| 43 |
248505_at |
AT5G50360
|
unknown |
0.856 |
-0.007 |
| 44 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.853 |
0.141 |
| 45 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.851 |
-0.045 |
| 46 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.845 |
0.068 |
| 47 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.833 |
-0.099 |
| 48 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.828 |
0.081 |
| 49 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.823 |
0.076 |
| 50 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.811 |
0.167 |
| 51 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
0.809 |
-0.344 |
| 52 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.804 |
0.005 |
| 53 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.794 |
0.233 |
| 54 |
264618_at |
AT2G17680
|
unknown |
0.792 |
0.107 |
| 55 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.779 |
0.126 |
| 56 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
0.771 |
-0.065 |
| 57 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
0.768 |
0.254 |
| 58 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.768 |
0.091 |
| 59 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.768 |
0.091 |
| 60 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.765 |
0.037 |
| 61 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.762 |
0.091 |
| 62 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.758 |
0.020 |
| 63 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.746 |
0.071 |
| 64 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.743 |
0.195 |
| 65 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.743 |
0.328 |
| 66 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.737 |
-0.017 |
| 67 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.730 |
-0.005 |
| 68 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.730 |
0.312 |
| 69 |
248424_at |
AT5G51680
|
[hydroxyproline-rich glycoprotein family protein] |
0.713 |
-0.039 |
| 70 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
0.712 |
0.134 |
| 71 |
255527_at |
AT4G02360
|
unknown |
0.711 |
-0.166 |
| 72 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
0.709 |
0.100 |
| 73 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.709 |
-0.261 |
| 74 |
247511_at |
AT5G62040
|
BFT, brother of FT and TFL1 |
0.702 |
-0.223 |
| 75 |
258878_at |
AT3G03170
|
unknown |
0.696 |
-0.019 |
| 76 |
249622_at |
AT5G37550
|
unknown |
0.683 |
0.044 |
| 77 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.678 |
0.368 |
| 78 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.668 |
-0.028 |
| 79 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.668 |
0.073 |
| 80 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.666 |
-0.062 |
| 81 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.654 |
-0.335 |
| 82 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.653 |
0.253 |
| 83 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.647 |
-0.023 |
| 84 |
253407_at |
AT4G32920
|
[glycine-rich protein] |
0.628 |
-0.099 |
| 85 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.620 |
0.159 |
| 86 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.611 |
-0.282 |
| 87 |
258091_at |
AT3G14560
|
unknown |
0.606 |
-0.149 |
| 88 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
0.601 |
-0.003 |
| 89 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
0.599 |
0.071 |
| 90 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
0.593 |
-0.013 |
| 91 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.590 |
0.100 |
| 92 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
0.571 |
0.117 |
| 93 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.568 |
-0.009 |
| 94 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.551 |
0.384 |
| 95 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.540 |
0.139 |
| 96 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-1.141 |
0.191 |
| 97 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-1.081 |
0.224 |
| 98 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.961 |
0.159 |
| 99 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.804 |
0.143 |
| 100 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.722 |
0.179 |
| 101 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.713 |
0.155 |
| 102 |
262211_at |
AT1G74930
|
ORA47 |
-0.711 |
0.138 |
| 103 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.687 |
0.017 |
| 104 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.629 |
0.006 |
| 105 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.619 |
-1.160 |
| 106 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.613 |
0.007 |
| 107 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.610 |
0.079 |
| 108 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.600 |
0.084 |
| 109 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.584 |
-0.010 |
| 110 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.578 |
0.145 |
| 111 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.540 |
-0.070 |
| 112 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
-0.515 |
0.130 |
| 113 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.511 |
-0.287 |
| 114 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.509 |
0.029 |
| 115 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.497 |
0.004 |
| 116 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.496 |
0.049 |
| 117 |
262286_at |
AT1G68585
|
unknown |
-0.492 |
-0.078 |
| 118 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.485 |
0.068 |
| 119 |
248942_at |
AT5G45480
|
unknown |
-0.480 |
-0.062 |
| 120 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.477 |
0.188 |
| 121 |
267367_at |
AT2G44210
|
unknown |
-0.467 |
-0.127 |
| 122 |
247183_at |
AT5G65440
|
unknown |
-0.446 |
-0.330 |
| 123 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.445 |
0.044 |
| 124 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.442 |
0.056 |
| 125 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
-0.434 |
0.235 |
| 126 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.433 |
0.093 |
| 127 |
259731_at |
AT1G77460
|
CSI3, CELLULOSE SYNTHASE INTERACTIVE 3 |
-0.432 |
-0.189 |
| 128 |
248186_at |
AT5G53880
|
unknown |
-0.422 |
0.062 |
| 129 |
249167_at |
AT5G42860
|
unknown |
-0.418 |
-0.263 |
| 130 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.410 |
0.206 |
| 131 |
252236_at |
AT3G49930
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.406 |
0.083 |
| 132 |
252446_at |
AT3G47010
|
[Glycosyl hydrolase family protein] |
-0.404 |
0.181 |
| 133 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.393 |
0.004 |
| 134 |
249378_at |
AT5G40450
|
unknown |
-0.392 |
-0.204 |
| 135 |
264609_at |
AT1G04530
|
TPR4, tetratricopeptide repeat 4 |
-0.391 |
-0.138 |
| 136 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.390 |
0.257 |
| 137 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
-0.388 |
0.617 |
| 138 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.387 |
-0.027 |
| 139 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.386 |
0.239 |
| 140 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.385 |
0.339 |
| 141 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.379 |
0.388 |
| 142 |
256547_at |
AT3G14840
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.377 |
0.107 |
| 143 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.375 |
0.248 |
| 144 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.369 |
-0.019 |
| 145 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.368 |
0.097 |
| 146 |
267637_at |
AT2G42190
|
unknown |
-0.367 |
-0.219 |
| 147 |
245937_at |
AT5G19750
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.362 |
-0.311 |
| 148 |
260395_at |
AT1G69780
|
ATHB13 |
-0.354 |
0.155 |
| 149 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
-0.352 |
-0.182 |
| 150 |
262442_at |
AT1G47420
|
SDH5, succinate dehydrogenase 5 |
-0.350 |
-0.019 |
| 151 |
259491_at |
AT1G15820
|
CP24, LHCB6, light harvesting complex photosystem II subunit 6 |
-0.341 |
0.138 |
| 152 |
247966_at |
AT5G56610
|
[Phosphotyrosine protein phosphatases superfamily protein] |
-0.338 |
-0.043 |
| 153 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.333 |
0.130 |
| 154 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
-0.331 |
0.201 |
| 155 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.327 |
0.323 |
| 156 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.325 |
0.098 |
| 157 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-0.324 |
0.009 |
| 158 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.324 |
-0.022 |
| 159 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.320 |
0.051 |
| 160 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.318 |
0.148 |
| 161 |
255277_at |
AT4G04890
|
PDF2, protodermal factor 2 |
-0.316 |
0.081 |
| 162 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.314 |
0.019 |
| 163 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.310 |
0.079 |
| 164 |
257510_at |
AT1G55360
|
unknown |
-0.306 |
-0.069 |
| 165 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.304 |
-0.080 |
| 166 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.301 |
0.038 |
| 167 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
-0.301 |
0.125 |
| 168 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.295 |
0.127 |
| 169 |
265327_at |
AT2G18210
|
unknown |
-0.294 |
0.217 |
| 170 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.287 |
0.123 |
| 171 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.287 |
0.171 |
| 172 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.286 |
-0.136 |
| 173 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.285 |
-0.029 |
| 174 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.285 |
0.246 |
| 175 |
259460_at |
AT1G44000
|
unknown |
-0.277 |
-0.347 |
| 176 |
250254_at |
AT5G13710
|
CPH, CEPHALOPOD, SMT1, sterol methyltransferase 1 |
-0.276 |
-0.196 |
| 177 |
245746_at |
AT1G51070
|
bHLH115, basic Helix-Loop-Helix 115 |
-0.275 |
-0.043 |
| 178 |
247567_at |
AT5G61190
|
[putative endonuclease or glycosyl hydrolase with C2H2-type zinc finger domain] |
-0.274 |
0.053 |
| 179 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.273 |
0.063 |
| 180 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.273 |
0.101 |
| 181 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.272 |
0.062 |
| 182 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
-0.272 |
0.157 |
| 183 |
248835_at |
AT5G47250
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.271 |
-0.005 |
| 184 |
264700_at |
AT1G70100
|
unknown |
-0.270 |
0.039 |
| 185 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.270 |
0.213 |
| 186 |
266338_at |
AT2G32400
|
ATGLR3.7, GLR3.7, GLUTAMATE RECEPTOR 3.7, GLR5, glutamate receptor 5 |
-0.269 |
0.092 |
| 187 |
249216_at |
AT5G42240
|
scpl42, serine carboxypeptidase-like 42 |
-0.268 |
0.276 |
| 188 |
256115_at |
AT1G16880
|
ACR11, ACT domain repeats 11 |
-0.266 |
0.094 |
| 189 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
-0.264 |
-0.085 |