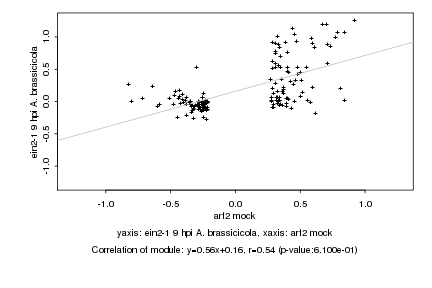
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
arf2 mock |
Log2 signal ratio
ein2-1 9 hpi A. brassicicola |
| 1 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.916 |
1.270 |
| 2 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.840 |
1.073 |
| 3 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.838 |
0.027 |
| 4 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.806 |
0.202 |
| 5 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.785 |
1.075 |
| 6 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.765 |
1.004 |
| 7 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.731 |
0.862 |
| 8 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.708 |
0.602 |
| 9 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.704 |
0.895 |
| 10 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.698 |
1.198 |
| 11 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.666 |
1.200 |
| 12 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.612 |
-0.179 |
| 13 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.608 |
0.838 |
| 14 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.591 |
0.910 |
| 15 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.588 |
0.222 |
| 16 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.580 |
0.985 |
| 17 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
0.573 |
-0.013 |
| 18 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.552 |
0.027 |
| 19 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.548 |
0.541 |
| 20 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.512 |
0.148 |
| 21 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.502 |
0.329 |
| 22 |
245041_at |
AT2G26530
|
AR781 |
0.501 |
0.464 |
| 23 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.500 |
0.079 |
| 24 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.477 |
0.528 |
| 25 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.471 |
0.420 |
| 26 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.470 |
0.933 |
| 27 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.459 |
0.328 |
| 28 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.454 |
0.006 |
| 29 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.449 |
1.042 |
| 30 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.445 |
0.265 |
| 31 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.433 |
1.132 |
| 32 |
245352_at |
AT4G15490
|
UGT84A3 |
0.430 |
-0.100 |
| 33 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.422 |
0.324 |
| 34 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.408 |
0.453 |
| 35 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.403 |
0.044 |
| 36 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.399 |
0.468 |
| 37 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.398 |
0.764 |
| 38 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.396 |
0.054 |
| 39 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.394 |
0.538 |
| 40 |
261542_at |
AT1G63560
|
[Receptor-like protein kinase-related family protein] |
0.391 |
-0.030 |
| 41 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.388 |
0.057 |
| 42 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.388 |
-0.064 |
| 43 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.380 |
0.924 |
| 44 |
260744_at |
AT1G15010
|
unknown |
0.369 |
0.130 |
| 45 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
0.368 |
0.232 |
| 46 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.364 |
0.192 |
| 47 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.362 |
-0.049 |
| 48 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
0.360 |
0.158 |
| 49 |
251847_at |
AT3G54640
|
TRP3, TRYPTOPHAN-REQUIRING 3, TSA1, tryptophan synthase alpha chain |
0.350 |
0.342 |
| 50 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.346 |
0.700 |
| 51 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.344 |
0.539 |
| 52 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.343 |
-0.042 |
| 53 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
0.337 |
0.031 |
| 54 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.336 |
0.068 |
| 55 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.336 |
0.027 |
| 56 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.335 |
0.851 |
| 57 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
0.334 |
-0.049 |
| 58 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.330 |
0.029 |
| 59 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
0.328 |
0.009 |
| 60 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.326 |
0.564 |
| 61 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.326 |
0.895 |
| 62 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
0.322 |
-0.029 |
| 63 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
0.321 |
0.160 |
| 64 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.319 |
0.072 |
| 65 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.318 |
1.012 |
| 66 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.315 |
0.063 |
| 67 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.311 |
0.026 |
| 68 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.308 |
0.750 |
| 69 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.308 |
0.901 |
| 70 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.306 |
0.291 |
| 71 |
260522_x_at |
AT2G41730
|
unknown |
0.304 |
0.779 |
| 72 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.302 |
0.589 |
| 73 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.302 |
0.542 |
| 74 |
252222_at |
AT3G49845
|
WIH3, WINDHOSE 3 |
0.293 |
0.128 |
| 75 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.289 |
-0.079 |
| 76 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
0.286 |
-0.039 |
| 77 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.284 |
0.620 |
| 78 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
0.282 |
0.216 |
| 79 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.281 |
0.929 |
| 80 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.280 |
-0.079 |
| 81 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.279 |
0.521 |
| 82 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.278 |
0.068 |
| 83 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.276 |
0.026 |
| 84 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.273 |
0.007 |
| 85 |
266167_at |
AT2G38860
|
DJ-1e, DJ1E, YLS5 |
0.268 |
0.349 |
| 86 |
259653_at |
AT1G55240
|
unknown |
-0.828 |
0.279 |
| 87 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.805 |
0.001 |
| 88 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.717 |
0.049 |
| 89 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.644 |
0.243 |
| 90 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.602 |
-0.077 |
| 91 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.586 |
-0.035 |
| 92 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.509 |
0.047 |
| 93 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.478 |
-0.037 |
| 94 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.477 |
0.107 |
| 95 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.470 |
0.167 |
| 96 |
248028_at |
AT5G55620
|
unknown |
-0.447 |
-0.235 |
| 97 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
-0.443 |
0.061 |
| 98 |
247882_at |
AT5G57785
|
unknown |
-0.439 |
0.082 |
| 99 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.432 |
0.184 |
| 100 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.424 |
-0.017 |
| 101 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.413 |
0.043 |
| 102 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.410 |
0.116 |
| 103 |
262121_at |
AT1G02800
|
ATCEL2, cellulase 2, CEL2, CEL2, cellulase 2 |
-0.408 |
-0.014 |
| 104 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.390 |
0.017 |
| 105 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.383 |
0.069 |
| 106 |
249010_at |
AT5G44580
|
unknown |
-0.383 |
-0.033 |
| 107 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.383 |
-0.209 |
| 108 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.356 |
-0.001 |
| 109 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.354 |
-0.037 |
| 110 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
-0.349 |
0.009 |
| 111 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
-0.348 |
0.003 |
| 112 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.341 |
-0.071 |
| 113 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.340 |
-0.170 |
| 114 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.338 |
-0.059 |
| 115 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.334 |
-0.132 |
| 116 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.326 |
-0.122 |
| 117 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.326 |
-0.258 |
| 118 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.326 |
-0.096 |
| 119 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.320 |
-0.064 |
| 120 |
265083_at |
AT1G03820
|
unknown |
-0.311 |
-0.050 |
| 121 |
245076_at |
AT2G23170
|
GH3.3 |
-0.305 |
0.533 |
| 122 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.297 |
-0.035 |
| 123 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.289 |
-0.113 |
| 124 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.288 |
0.001 |
| 125 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
-0.287 |
-0.045 |
| 126 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.287 |
-0.077 |
| 127 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
-0.284 |
-0.011 |
| 128 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.283 |
-0.079 |
| 129 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.282 |
-0.077 |
| 130 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
-0.272 |
-0.118 |
| 131 |
246860_at |
AT5G25840
|
unknown |
-0.265 |
-0.031 |
| 132 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.264 |
-0.153 |
| 133 |
263528_at |
AT2G24800
|
[Peroxidase superfamily protein] |
-0.260 |
0.070 |
| 134 |
246877_at |
AT5G26150
|
[protein kinase family protein] |
-0.260 |
-0.015 |
| 135 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.259 |
0.063 |
| 136 |
246487_at |
AT5G16030
|
unknown |
-0.259 |
-0.082 |
| 137 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.258 |
-0.110 |
| 138 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.258 |
0.007 |
| 139 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.256 |
-0.082 |
| 140 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.256 |
-0.084 |
| 141 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.255 |
-0.012 |
| 142 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.252 |
-0.244 |
| 143 |
258530_at |
AT3G06840
|
unknown |
-0.251 |
-0.089 |
| 144 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.250 |
-0.135 |
| 145 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.248 |
-0.055 |
| 146 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.247 |
0.126 |
| 147 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.241 |
-0.016 |
| 148 |
248245_at |
AT5G53190
|
AtSWEET3, SWEET3 |
-0.240 |
-0.069 |
| 149 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
-0.238 |
-0.029 |
| 150 |
262260_at |
AT1G70850
|
MLP34, MLP-like protein 34 |
-0.235 |
0.029 |
| 151 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.234 |
-0.097 |
| 152 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.234 |
-0.001 |
| 153 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
-0.234 |
-0.095 |
| 154 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.231 |
-0.014 |
| 155 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.231 |
-0.026 |
| 156 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.228 |
-0.131 |
| 157 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.228 |
-0.139 |
| 158 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.226 |
-0.112 |
| 159 |
266179_at |
AT2G02300
|
AtPP2-B5, phloem protein 2-B5, PP2-B5, phloem protein 2-B5 |
-0.225 |
0.004 |
| 160 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.224 |
-0.263 |
| 161 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
-0.224 |
-0.024 |
| 162 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.222 |
-0.012 |
| 163 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.221 |
0.014 |
| 164 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.218 |
0.008 |
| 165 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.218 |
-0.003 |
| 166 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.217 |
-0.121 |
| 167 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.216 |
-0.119 |
| 168 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
-0.216 |
-0.088 |
| 169 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
-0.216 |
-0.006 |