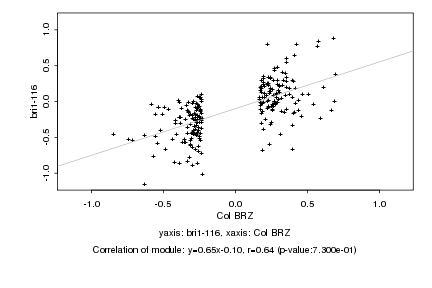
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col BRZ |
Log2 signal ratio
bri1-116 |
| 1 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.690 |
0.389 |
| 2 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
0.687 |
0.012 |
| 3 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.678 |
0.885 |
| 4 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
0.667 |
-0.121 |
| 5 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.607 |
0.205 |
| 6 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.589 |
-0.235 |
| 7 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.572 |
0.850 |
| 8 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.567 |
0.773 |
| 9 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.540 |
-0.041 |
| 10 |
244994_at |
ATCG01010
|
NDHF |
0.504 |
0.104 |
| 11 |
261777_at |
AT1G76210
|
unknown |
0.465 |
0.106 |
| 12 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.453 |
-0.198 |
| 13 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
0.438 |
-0.114 |
| 14 |
257334_at |
ATMG01370
|
ORF111D |
0.437 |
0.020 |
| 15 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.423 |
0.799 |
| 16 |
264774_at |
AT1G22890
|
unknown |
0.412 |
0.187 |
| 17 |
252571_at |
AT3G45280
|
ATSYP72, SYP72, syntaxin of plants 72 |
0.411 |
-0.026 |
| 18 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.407 |
-0.151 |
| 19 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.406 |
0.647 |
| 20 |
248028_at |
AT5G55620
|
unknown |
0.403 |
0.291 |
| 21 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.399 |
-0.166 |
| 22 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
0.393 |
0.064 |
| 23 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.392 |
0.306 |
| 24 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.390 |
-0.332 |
| 25 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.390 |
-0.657 |
| 26 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.372 |
0.185 |
| 27 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.371 |
0.099 |
| 28 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
0.354 |
-0.104 |
| 29 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
0.353 |
0.610 |
| 30 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
0.351 |
0.289 |
| 31 |
249482_at |
AT5G38980
|
unknown |
0.350 |
0.203 |
| 32 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.350 |
0.345 |
| 33 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.348 |
0.548 |
| 34 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
0.346 |
0.392 |
| 35 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.341 |
0.089 |
| 36 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.334 |
-0.152 |
| 37 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
0.327 |
0.300 |
| 38 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.326 |
0.405 |
| 39 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
0.324 |
0.235 |
| 40 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
0.316 |
-0.131 |
| 41 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
0.314 |
0.047 |
| 42 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
0.313 |
-0.455 |
| 43 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.306 |
0.220 |
| 44 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
0.302 |
0.133 |
| 45 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.301 |
0.236 |
| 46 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
0.299 |
0.160 |
| 47 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
0.293 |
0.031 |
| 48 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.289 |
0.239 |
| 49 |
246842_at |
AT5G26731
|
unknown |
0.289 |
-0.009 |
| 50 |
262981_at |
AT1G75590
|
SAUR52, SMALL AUXIN UPREGULATED RNA 52 |
0.287 |
0.476 |
| 51 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.287 |
0.307 |
| 52 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.285 |
0.057 |
| 53 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.283 |
0.108 |
| 54 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
0.279 |
0.114 |
| 55 |
266877_at |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
0.274 |
-0.022 |
| 56 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.274 |
-0.031 |
| 57 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
0.271 |
0.009 |
| 58 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
0.271 |
0.440 |
| 59 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
0.271 |
0.014 |
| 60 |
266814_at |
AT2G44910
|
ATHB-4, ARABIDOPSIS THALIANA HOMEOBOX-LEUCINE ZIPPER PROTEIN 4, ATHB4, homeobox-leucine zipper protein 4, HB4, homeobox-leucine zipper protein 4 |
0.268 |
0.467 |
| 61 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
0.264 |
-0.061 |
| 62 |
248584_at |
AT5G49960
|
unknown |
0.260 |
0.097 |
| 63 |
257357_at |
AT2G41050
|
[PQ-loop repeat family protein / transmembrane family protein] |
0.259 |
-0.031 |
| 64 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.258 |
0.302 |
| 65 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.257 |
-0.104 |
| 66 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.257 |
0.179 |
| 67 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
0.255 |
-0.036 |
| 68 |
248230_at |
AT5G53830
|
[VQ motif-containing protein] |
0.251 |
-0.117 |
| 69 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.248 |
-0.066 |
| 70 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
0.244 |
-0.280 |
| 71 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.243 |
0.325 |
| 72 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.239 |
0.336 |
| 73 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
0.239 |
-0.311 |
| 74 |
263237_at |
AT1G10610
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.239 |
0.146 |
| 75 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
0.237 |
0.193 |
| 76 |
263586_at |
AT2G25350
|
[Phox (PX) domain-containing protein] |
0.233 |
0.141 |
| 77 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.232 |
-0.598 |
| 78 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
0.230 |
0.119 |
| 79 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.229 |
0.018 |
| 80 |
253813_at |
AT4G28150
|
unknown |
0.226 |
0.264 |
| 81 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.225 |
0.342 |
| 82 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.224 |
0.228 |
| 83 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.224 |
-0.080 |
| 84 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
0.223 |
-0.062 |
| 85 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.222 |
0.004 |
| 86 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.218 |
0.801 |
| 87 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.218 |
-0.085 |
| 88 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.216 |
0.247 |
| 89 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.216 |
0.265 |
| 90 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.209 |
0.094 |
| 91 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.204 |
-0.239 |
| 92 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
0.201 |
0.124 |
| 93 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
0.199 |
0.222 |
| 94 |
253297_at |
AT4G33810
|
[Glycosyl hydrolase superfamily protein] |
0.199 |
0.069 |
| 95 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
0.194 |
-0.387 |
| 96 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
0.194 |
0.277 |
| 97 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.192 |
0.238 |
| 98 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
0.192 |
-0.007 |
| 99 |
263971_at |
AT2G42800
|
AtRLP29, receptor like protein 29, RLP29, receptor like protein 29 |
0.189 |
0.333 |
| 100 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.189 |
0.354 |
| 101 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.186 |
-0.025 |
| 102 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.184 |
-0.683 |
| 103 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.183 |
-0.131 |
| 104 |
257375_at |
AT2G38640
|
unknown |
0.183 |
0.064 |
| 105 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
0.181 |
-0.162 |
| 106 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.180 |
0.303 |
| 107 |
248140_at |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
0.180 |
0.130 |
| 108 |
263002_at |
AT1G54200
|
unknown |
0.177 |
-0.020 |
| 109 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
0.177 |
-0.294 |
| 110 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
0.177 |
0.191 |
| 111 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
0.176 |
0.210 |
| 112 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.174 |
-0.111 |
| 113 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
0.174 |
-0.013 |
| 114 |
261456_at |
AT1G21050
|
unknown |
0.174 |
0.201 |
| 115 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
0.173 |
-0.049 |
| 116 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
0.169 |
0.157 |
| 117 |
263700_at |
AT1G31150
|
[FUNCTIONS IN: sequence-specific DNA binding transcription factor activity] |
0.167 |
0.060 |
| 118 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.166 |
0.037 |
| 119 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.848 |
-0.459 |
| 120 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.744 |
-0.529 |
| 121 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
-0.722 |
-0.541 |
| 122 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.633 |
-0.461 |
| 123 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.632 |
-1.144 |
| 124 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.586 |
-0.029 |
| 125 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.575 |
-0.759 |
| 126 |
265083_at |
AT1G03820
|
unknown |
-0.562 |
-0.173 |
| 127 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.557 |
-0.479 |
| 128 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.543 |
-0.575 |
| 129 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.537 |
-0.073 |
| 130 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-0.535 |
-0.075 |
| 131 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-0.524 |
-0.394 |
| 132 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.508 |
-0.176 |
| 133 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-0.495 |
-0.070 |
| 134 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.491 |
-0.667 |
| 135 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.472 |
-0.111 |
| 136 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.441 |
-0.519 |
| 137 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.426 |
-0.843 |
| 138 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.422 |
-0.293 |
| 139 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
-0.418 |
-0.263 |
| 140 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.416 |
-0.452 |
| 141 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
-0.401 |
0.019 |
| 142 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
-0.394 |
-0.013 |
| 143 |
250472_at |
AT5G10210
|
unknown |
-0.391 |
-0.210 |
| 144 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-0.389 |
-0.221 |
| 145 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.389 |
-0.854 |
| 146 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
-0.387 |
-0.304 |
| 147 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
-0.384 |
-0.218 |
| 148 |
246415_at |
AT5G17160
|
unknown |
-0.376 |
-0.097 |
| 149 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.371 |
-0.566 |
| 150 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.359 |
-0.524 |
| 151 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
-0.358 |
-0.561 |
| 152 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.353 |
-0.565 |
| 153 |
260332_at |
AT1G70470
|
unknown |
-0.352 |
-0.242 |
| 154 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.342 |
-0.047 |
| 155 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.340 |
-0.828 |
| 156 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.336 |
-0.352 |
| 157 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.335 |
-0.117 |
| 158 |
267618_at |
AT2G26760
|
CYCB1;4, Cyclin B1;4 |
-0.334 |
-0.445 |
| 159 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.330 |
-0.141 |
| 160 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.327 |
-0.000 |
| 161 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
-0.327 |
-0.132 |
| 162 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.324 |
-0.768 |
| 163 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-0.324 |
-0.184 |
| 164 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.316 |
-0.601 |
| 165 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
-0.315 |
-0.316 |
| 166 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.315 |
-0.594 |
| 167 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
-0.314 |
-0.176 |
| 168 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
-0.313 |
-0.537 |
| 169 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.311 |
-0.442 |
| 170 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.308 |
-0.507 |
| 171 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
-0.304 |
-0.024 |
| 172 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.304 |
-0.890 |
| 173 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.303 |
-0.425 |
| 174 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.302 |
-0.190 |
| 175 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.299 |
-0.640 |
| 176 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
-0.299 |
-0.227 |
| 177 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.296 |
0.006 |
| 178 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.296 |
-0.237 |
| 179 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.295 |
-0.402 |
| 180 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.292 |
-0.448 |
| 181 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
-0.291 |
-0.165 |
| 182 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.290 |
-0.339 |
| 183 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.286 |
-0.433 |
| 184 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.286 |
-0.335 |
| 185 |
259962_at |
AT1G53690
|
[DNA directed RNA polymerase, 7 kDa subunit] |
-0.284 |
-0.664 |
| 186 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.284 |
-0.258 |
| 187 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.283 |
-0.075 |
| 188 |
253100_at |
AT4G37400
|
CYP81F3, cytochrome P450, family 81, subfamily F, polypeptide 3 |
-0.283 |
-0.169 |
| 189 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-0.283 |
-0.257 |
| 190 |
262121_at |
AT1G02800
|
ATCEL2, cellulase 2, CEL2, CEL2, cellulase 2 |
-0.282 |
-0.253 |
| 191 |
265327_at |
AT2G18210
|
unknown |
-0.279 |
-0.381 |
| 192 |
245524_at |
AT4G15920
|
AtSWEET17, SWEET17 |
-0.278 |
-0.222 |
| 193 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
-0.277 |
-0.324 |
| 194 |
260522_x_at |
AT2G41730
|
unknown |
-0.275 |
-0.458 |
| 195 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.273 |
-0.014 |
| 196 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
-0.273 |
-0.291 |
| 197 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.273 |
-0.118 |
| 198 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.272 |
-0.464 |
| 199 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
-0.270 |
0.074 |
| 200 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.269 |
-0.244 |
| 201 |
247425_at |
AT5G62550
|
unknown |
-0.269 |
-0.095 |
| 202 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
-0.267 |
-0.185 |
| 203 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.267 |
-0.861 |
| 204 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.266 |
-0.286 |
| 205 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.265 |
-0.431 |
| 206 |
258873_at |
AT3G03240
|
[alpha/beta-Hydrolases superfamily protein] |
-0.264 |
-0.041 |
| 207 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.263 |
-0.392 |
| 208 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.262 |
-0.360 |
| 209 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.262 |
-0.618 |
| 210 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.262 |
-0.691 |
| 211 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.261 |
-0.069 |
| 212 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.255 |
-0.222 |
| 213 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.254 |
0.070 |
| 214 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
-0.252 |
-0.058 |
| 215 |
250375_at |
AT5G11540
|
AtGulLO3, GulLO3, L -gulono-1,4-lactone ( L -GulL) oxidase 3 |
-0.251 |
-0.290 |
| 216 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.250 |
-0.203 |
| 217 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
-0.250 |
-0.507 |
| 218 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.250 |
-0.484 |
| 219 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.249 |
-0.101 |
| 220 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
-0.249 |
-0.130 |
| 221 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.248 |
0.057 |
| 222 |
259290_at |
AT3G11520
|
CYC2, CYCLIN 2, CYCB1;3, CYCLIN B1;3 |
-0.245 |
-0.438 |
| 223 |
263515_at |
AT2G21640
|
unknown |
-0.243 |
-0.118 |
| 224 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.243 |
-0.531 |
| 225 |
258480_at |
AT3G02640
|
unknown |
-0.243 |
-0.243 |
| 226 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.242 |
-0.715 |
| 227 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
-0.241 |
-0.369 |
| 228 |
253305_at |
AT4G33666
|
unknown |
-0.241 |
-0.290 |
| 229 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.240 |
0.036 |
| 230 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.240 |
-0.157 |
| 231 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.239 |
0.008 |
| 232 |
260795_at |
AT1G06225
|
CLE3, CLAVATA3/ESR-RELATED 3 |
-0.237 |
0.101 |
| 233 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.236 |
-0.263 |
| 234 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
-0.235 |
-0.231 |
| 235 |
264341_at |
AT1G70270
|
unknown |
-0.234 |
-1.016 |