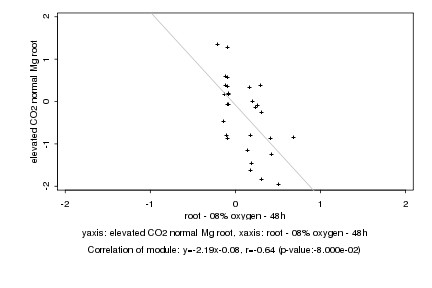
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root - 08% oxygen - 48h |
Log2 signal ratio
elevated CO2 normal Mg root |
| 1 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.680 |
-0.826 |
| 2 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.501 |
-1.935 |
| 3 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.414 |
-1.249 |
| 4 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.406 |
-0.866 |
| 5 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.306 |
-1.818 |
| 6 |
250152_at |
AT5G15120
|
unknown |
0.304 |
-0.250 |
| 7 |
245026_at |
ATCG00140
|
ATPH |
0.286 |
0.389 |
| 8 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.258 |
-0.075 |
| 9 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.224 |
-0.135 |
| 10 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.192 |
0.010 |
| 11 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.177 |
-1.450 |
| 12 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.173 |
-1.606 |
| 13 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.170 |
-0.800 |
| 14 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.154 |
0.342 |
| 15 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.139 |
-1.154 |
| 16 |
258068_at |
AT3G25990
|
[Homeodomain-like superfamily protein] |
-0.211 |
1.347 |
| 17 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.149 |
-0.465 |
| 18 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
-0.129 |
0.179 |
| 19 |
254927_at |
AT4G11400
|
[ARID/BRIGHT DNA-binding domain] |
-0.124 |
0.383 |
| 20 |
250837_at |
AT5G04620
|
ATBIOF, biotin F, BIO4, biotin 4, BIOF, biotin F |
-0.123 |
0.610 |
| 21 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
-0.106 |
-0.790 |
| 22 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.105 |
0.365 |
| 23 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
-0.101 |
-0.054 |
| 24 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.098 |
0.572 |
| 25 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.098 |
1.287 |
| 26 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.098 |
-0.859 |
| 27 |
254670_at |
AT4G18390
|
TCP2, TCP2, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 |
-0.091 |
0.179 |
| 28 |
259296_at |
AT3G05350
|
[Metallopeptidase M24 family protein] |
-0.086 |
0.209 |
| 29 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.084 |
-0.054 |