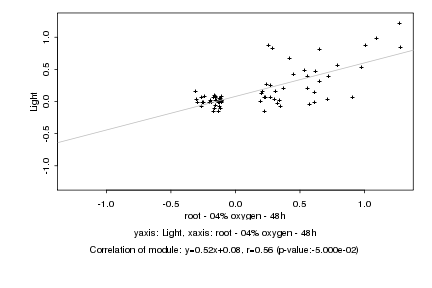
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root - 04% oxygen - 48h |
Log2 signal ratio
Light |
| 1 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.276 |
0.848 |
| 2 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.266 |
1.215 |
| 3 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.091 |
0.995 |
| 4 |
245226_at |
AT3G29970
|
[B12D protein] |
1.007 |
0.878 |
| 5 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.976 |
0.539 |
| 6 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.905 |
0.069 |
| 7 |
254200_at |
AT4G24110
|
unknown |
0.785 |
0.570 |
| 8 |
249384_at |
AT5G39890
|
unknown |
0.714 |
0.404 |
| 9 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.706 |
0.042 |
| 10 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.648 |
0.317 |
| 11 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.645 |
0.815 |
| 12 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.618 |
0.481 |
| 13 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.610 |
-0.014 |
| 14 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.607 |
0.141 |
| 15 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.573 |
-0.033 |
| 16 |
250152_at |
AT5G15120
|
unknown |
0.553 |
0.213 |
| 17 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.551 |
0.397 |
| 18 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.533 |
0.491 |
| 19 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.447 |
0.434 |
| 20 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.417 |
0.684 |
| 21 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.365 |
0.215 |
| 22 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.345 |
-0.077 |
| 23 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.339 |
0.029 |
| 24 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.323 |
-0.026 |
| 25 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.308 |
0.167 |
| 26 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.296 |
0.037 |
| 27 |
261567_at |
AT1G33055
|
unknown |
0.282 |
0.837 |
| 28 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.269 |
0.258 |
| 29 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.266 |
0.071 |
| 30 |
247024_at |
AT5G66985
|
unknown |
0.249 |
0.876 |
| 31 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.234 |
0.269 |
| 32 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.226 |
0.069 |
| 33 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.224 |
-0.148 |
| 34 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.224 |
0.063 |
| 35 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.209 |
0.157 |
| 36 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.195 |
0.139 |
| 37 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
0.192 |
0.002 |
| 38 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.317 |
0.160 |
| 39 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.302 |
0.041 |
| 40 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.296 |
-0.006 |
| 41 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.268 |
0.066 |
| 42 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.264 |
-0.067 |
| 43 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.263 |
-0.001 |
| 44 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
-0.248 |
-0.008 |
| 45 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
-0.246 |
0.090 |
| 46 |
267471_at |
AT2G30390
|
ATFC-II, FC-II, FC2, ferrochelatase 2 |
-0.208 |
-0.000 |
| 47 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
-0.195 |
0.023 |
| 48 |
260599_at |
AT1G55940
|
CYP708A1, cytochrome P450, family 708, subfamily A, polypeptide 1 |
-0.191 |
-0.009 |
| 49 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
-0.175 |
0.073 |
| 50 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.171 |
-0.150 |
| 51 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.164 |
-0.107 |
| 52 |
253717_at |
AT4G29440
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.163 |
0.096 |
| 53 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.161 |
0.029 |
| 54 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.160 |
-0.048 |
| 55 |
267296_at |
AT2G23660
|
LBD10, LOB domain-containing protein 10 |
-0.156 |
0.090 |
| 56 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.155 |
0.068 |
| 57 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.152 |
0.018 |
| 58 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.135 |
-0.011 |
| 59 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.134 |
0.009 |
| 60 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.133 |
-0.151 |
| 61 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
-0.131 |
0.059 |
| 62 |
255338_at |
AT4G04470
|
PMP22 |
-0.130 |
-0.077 |
| 63 |
254155_at |
AT4G24480
|
[Protein kinase superfamily protein] |
-0.127 |
-0.012 |
| 64 |
263811_at |
AT2G04350
|
LACS8, long-chain acyl-CoA synthetase 8 |
-0.126 |
-0.008 |
| 65 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
-0.125 |
0.005 |
| 66 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.121 |
-0.105 |
| 67 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
-0.117 |
0.049 |
| 68 |
244904_at |
ATMG00670
|
ORF275 |
-0.117 |
0.020 |
| 69 |
258944_at |
AT3G10390
|
FLD, FLOWERING LOCUS D |
-0.115 |
0.022 |
| 70 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-0.115 |
0.022 |
| 71 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.115 |
0.009 |
| 72 |
247967_at |
AT5G56620
|
anac099, NAC domain containing protein 99, NAC099, NAC domain containing protein 99 |
-0.114 |
-0.005 |
| 73 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.114 |
0.083 |