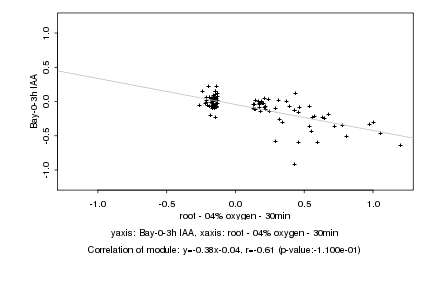
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root - 04% oxygen - 30min |
Log2 signal ratio
Bay-0-3h IAA |
| 1 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.197 |
-0.638 |
| 2 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.050 |
-0.460 |
| 3 |
250464_at |
AT5G10040
|
unknown |
1.002 |
-0.306 |
| 4 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.970 |
-0.327 |
| 5 |
254200_at |
AT4G24110
|
unknown |
0.803 |
-0.499 |
| 6 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.778 |
-0.339 |
| 7 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.719 |
-0.357 |
| 8 |
249384_at |
AT5G39890
|
unknown |
0.673 |
-0.183 |
| 9 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.645 |
-0.234 |
| 10 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.632 |
-0.230 |
| 11 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.594 |
-0.598 |
| 12 |
250152_at |
AT5G15120
|
unknown |
0.571 |
-0.205 |
| 13 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.557 |
-0.227 |
| 14 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.551 |
-0.431 |
| 15 |
257925_at |
AT3G23170
|
unknown |
0.538 |
-0.059 |
| 16 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.535 |
-0.363 |
| 17 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.465 |
-0.085 |
| 18 |
261567_at |
AT1G33055
|
unknown |
0.457 |
-0.593 |
| 19 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.456 |
-0.159 |
| 20 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.435 |
0.122 |
| 21 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.428 |
-0.122 |
| 22 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.425 |
-0.907 |
| 23 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.424 |
-0.126 |
| 24 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.386 |
-0.060 |
| 25 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.368 |
0.012 |
| 26 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.336 |
-0.296 |
| 27 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.319 |
-0.251 |
| 28 |
245401_at |
AT4G17670
|
unknown |
0.312 |
0.017 |
| 29 |
254522_at |
AT4G19980
|
unknown |
0.288 |
-0.096 |
| 30 |
247024_at |
AT5G66985
|
unknown |
0.284 |
-0.574 |
| 31 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.242 |
-0.142 |
| 32 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.235 |
0.032 |
| 33 |
263850_at |
AT2G04480
|
unknown |
0.216 |
-0.071 |
| 34 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.215 |
-0.114 |
| 35 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
0.210 |
0.045 |
| 36 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.208 |
-0.084 |
| 37 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.202 |
-0.026 |
| 38 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.196 |
-0.024 |
| 39 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
0.189 |
0.005 |
| 40 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
0.178 |
-0.025 |
| 41 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.175 |
-0.144 |
| 42 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
0.171 |
-0.038 |
| 43 |
259685_at |
AT1G63090
|
AtPP2-A11, phloem protein 2-A11, PP2-A11, phloem protein 2-A11 |
0.171 |
-0.073 |
| 44 |
253965_at |
AT4G26490
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.168 |
-0.019 |
| 45 |
253017_at |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
0.164 |
0.012 |
| 46 |
267459_at |
AT2G33850
|
unknown |
0.145 |
-0.109 |
| 47 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.144 |
0.028 |
| 48 |
265247_at |
AT2G43030
|
[Ribosomal protein L3 family protein] |
0.135 |
-0.033 |
| 49 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.133 |
-0.109 |
| 50 |
250223_at |
AT5G14070
|
ROXY2 |
0.128 |
-0.034 |
| 51 |
259303_at |
AT3G05130
|
unknown |
0.125 |
-0.099 |
| 52 |
249440_at |
AT5G40030
|
[Protein kinase superfamily protein] |
-0.268 |
-0.052 |
| 53 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
-0.240 |
0.147 |
| 54 |
261055_at |
AT1G01300
|
[Eukaryotic aspartyl protease family protein] |
-0.220 |
-0.020 |
| 55 |
245166_at |
AT2G33170
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.217 |
0.064 |
| 56 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
-0.216 |
0.018 |
| 57 |
265840_at |
AT2G14530
|
TBL13, TRICHOME BIREFRINGENCE-LIKE 13 |
-0.212 |
-0.011 |
| 58 |
252047_at |
AT3G52490
|
SMXL3, SMAX1-like 3 |
-0.204 |
-0.054 |
| 59 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.201 |
0.233 |
| 60 |
250132_at |
AT5G16560
|
KAN, KANADI, KAN1, KANADI 1 |
-0.195 |
-0.070 |
| 61 |
254966_at |
AT4G11110
|
SPA2, SPA1-related 2 |
-0.193 |
0.065 |
| 62 |
260363_at |
AT1G70550
|
unknown |
-0.192 |
0.059 |
| 63 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.187 |
-0.190 |
| 64 |
257201_at |
AT3G23740
|
unknown |
-0.180 |
-0.065 |
| 65 |
247828_at |
AT5G58510
|
unknown |
-0.180 |
0.040 |
| 66 |
262797_at |
AT1G20840
|
AtTMT1, TMT1, tonoplast monosaccharide transporter1 |
-0.180 |
-0.008 |
| 67 |
246285_at |
AT4G36980
|
unknown |
-0.177 |
-0.003 |
| 68 |
258206_at |
AT3G14010
|
CID4, CTC-interacting domain 4 |
-0.177 |
0.051 |
| 69 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.174 |
0.053 |
| 70 |
247134_at |
AT5G66230
|
[Chalcone-flavanone isomerase family protein] |
-0.171 |
-0.052 |
| 71 |
262052_at |
AT1G80020
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 5.0e-62 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.171 |
-0.016 |
| 72 |
254206_at |
AT4G24180
|
ATTLP1, TLP1, THAUMATIN-LIKE PROTEIN 1 |
-0.170 |
-0.096 |
| 73 |
264980_at |
AT1G27190
|
[Leucine-rich repeat protein kinase family protein] |
-0.170 |
0.020 |
| 74 |
248438_at |
AT5G51230
|
AtEMF2, CYR1, EMF2, EMBRYONIC FLOWER 2, VEF2, CYTOKININ RESISTANT 1 |
-0.168 |
0.013 |
| 75 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
-0.165 |
0.089 |
| 76 |
252608_at |
AT3G45090
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.165 |
-0.078 |
| 77 |
250571_at |
AT5G08200
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.165 |
0.006 |
| 78 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.163 |
-0.036 |
| 79 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.162 |
0.009 |
| 80 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.159 |
0.032 |
| 81 |
249146_at |
AT5G43310
|
[COP1-interacting protein-related] |
-0.158 |
-0.054 |
| 82 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.158 |
-0.094 |
| 83 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.157 |
0.059 |
| 84 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
-0.153 |
0.018 |
| 85 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.150 |
-0.221 |
| 86 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.148 |
0.107 |
| 87 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.147 |
0.150 |
| 88 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.146 |
-0.044 |
| 89 |
250774_at |
AT5G05450
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.145 |
-0.034 |
| 90 |
255828_at |
AT2G40630
|
[Uncharacterised conserved protein (UCP030365)] |
-0.145 |
0.052 |
| 91 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.145 |
0.151 |
| 92 |
266444_at |
AT2G43150
|
[Proline-rich extensin-like family protein] |
-0.144 |
-0.060 |
| 93 |
249235_at |
AT5G42100
|
ATBG_PPAP, ARABIDOPSIS THALIANA BETA-1,3-GLUCANASE_PUTATIVE, BG_PPAP, beta-1,3-glucanase_putative |
-0.142 |
-0.077 |
| 94 |
253300_at |
AT4G33580
|
ATBCA5, A. THALIANA BETA CARBONIC ANHYDRASE 5, BCA5, beta carbonic anhydrase 5 |
-0.142 |
0.050 |
| 95 |
255788_at |
AT2G33310
|
IAA13, auxin-induced protein 13 |
-0.140 |
0.230 |
| 96 |
246801_at |
AT5G26830
|
[Threonyl-tRNA synthetase] |
-0.140 |
-0.065 |
| 97 |
247955_at |
AT5G56950
|
NAP1;3, nucleosome assembly protein 1;3, NFA03, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 03, NFA3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A3 |
-0.139 |
-0.022 |
| 98 |
247845_at |
AT5G58090
|
[O-Glycosyl hydrolases family 17 protein] |
-0.137 |
-0.026 |
| 99 |
265827_at |
AT2G42400
|
ATVOZ2, VASCULAR PLANT ONE ZINC FINGER PROTEIN 2, VOZ2, vascular plant one zinc finger protein 2 |
-0.137 |
0.078 |
| 100 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
-0.137 |
0.020 |
| 101 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.137 |
0.126 |