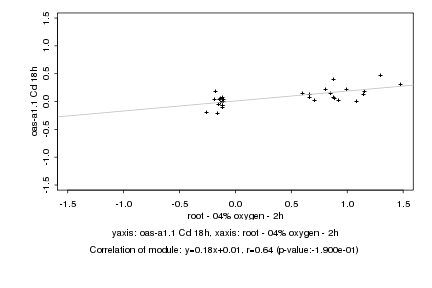
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root - 04% oxygen - 2h |
Log2 signal ratio
oas-a1.1 Cd 18h |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.473 |
0.312 |
| 2 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.291 |
0.481 |
| 3 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.151 |
0.197 |
| 4 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.141 |
0.144 |
| 5 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.074 |
0.005 |
| 6 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.985 |
0.230 |
| 7 |
254200_at |
AT4G24110
|
unknown |
0.917 |
0.026 |
| 8 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.879 |
0.068 |
| 9 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.873 |
0.412 |
| 10 |
249384_at |
AT5G39890
|
unknown |
0.873 |
0.080 |
| 11 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.842 |
0.158 |
| 12 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.802 |
0.224 |
| 13 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.706 |
0.024 |
| 14 |
250152_at |
AT5G15120
|
unknown |
0.657 |
0.088 |
| 15 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.655 |
0.143 |
| 16 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.593 |
0.145 |
| 17 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.260 |
-0.188 |
| 18 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.191 |
0.042 |
| 19 |
257056_at |
AT3G15350
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.179 |
0.192 |
| 20 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.169 |
-0.214 |
| 21 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
-0.158 |
-0.040 |
| 22 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.147 |
0.051 |
| 23 |
257512_at |
AT1G35250
|
ALT2, acyl-lipid thioesterase 2 |
-0.142 |
-0.003 |
| 24 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.141 |
0.059 |
| 25 |
248777_at |
AT5G47920
|
unknown |
-0.121 |
-0.096 |
| 26 |
265357_at |
AT2G16740
|
UBC29, ubiquitin-conjugating enzyme 29 |
-0.121 |
0.002 |
| 27 |
259220_at |
AT3G03550
|
[RING/U-box superfamily protein] |
-0.118 |
-0.066 |
| 28 |
263355_at |
AT2G22100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.117 |
-0.054 |
| 29 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.117 |
0.087 |
| 30 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.113 |
0.037 |
| 31 |
252134_at |
AT3G50910
|
unknown |
-0.110 |
0.011 |