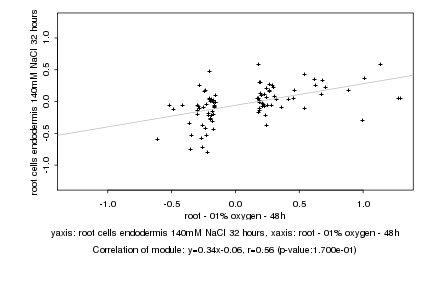
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root - 01% oxygen - 48h |
Log2 signal ratio
root cells endodermis 140mM NaCl 32 hours |
| 1 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.290 |
0.058 |
| 2 |
245226_at |
AT3G29970
|
[B12D protein] |
1.276 |
0.056 |
| 3 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.132 |
0.586 |
| 4 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.005 |
0.371 |
| 5 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.990 |
-0.286 |
| 6 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.878 |
0.188 |
| 7 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.699 |
0.232 |
| 8 |
254200_at |
AT4G24110
|
unknown |
0.678 |
0.331 |
| 9 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.667 |
0.126 |
| 10 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.626 |
0.264 |
| 11 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.619 |
0.359 |
| 12 |
250152_at |
AT5G15120
|
unknown |
0.536 |
0.436 |
| 13 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.534 |
-0.103 |
| 14 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.458 |
0.178 |
| 15 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.454 |
0.057 |
| 16 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.412 |
0.043 |
| 17 |
263515_at |
AT2G21640
|
unknown |
0.358 |
-0.085 |
| 18 |
256114_at |
AT1G16850
|
unknown |
0.318 |
0.034 |
| 19 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.302 |
0.087 |
| 20 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.298 |
0.221 |
| 21 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.288 |
0.266 |
| 22 |
266458_at |
AT2G47710
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.282 |
-0.052 |
| 23 |
264176_at |
AT1G02110
|
unknown |
0.262 |
0.188 |
| 24 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.262 |
0.282 |
| 25 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.259 |
0.159 |
| 26 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.244 |
-0.049 |
| 27 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.240 |
0.206 |
| 28 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.239 |
0.076 |
| 29 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.238 |
-0.377 |
| 30 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.228 |
-0.212 |
| 31 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.227 |
0.115 |
| 32 |
256578_at |
AT3G28200
|
[Peroxidase superfamily protein] |
0.222 |
-0.066 |
| 33 |
251958_at |
AT3G53560
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.216 |
-0.044 |
| 34 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.211 |
-0.075 |
| 35 |
260795_at |
AT1G06225
|
CLE3, CLAVATA3/ESR-RELATED 3 |
0.205 |
-0.019 |
| 36 |
247247_at |
AT5G64650
|
[Ribosomal protein L17 family protein] |
0.200 |
0.110 |
| 37 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.191 |
0.134 |
| 38 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.189 |
0.311 |
| 39 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.186 |
0.305 |
| 40 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.185 |
-0.013 |
| 41 |
251585_at |
AT3G58630
|
[sequence-specific DNA binding transcription factors] |
0.185 |
-0.105 |
| 42 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.181 |
-0.136 |
| 43 |
258987_at |
AT3G08950
|
HCC1, homologue of the copper chaperone SCO1 |
0.181 |
0.019 |
| 44 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.179 |
0.052 |
| 45 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.175 |
-0.157 |
| 46 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.173 |
0.592 |
| 47 |
266234_at |
AT2G02350
|
AtPP2-B9, Phloem protein 2-B9, SKIP3, SKP1 interacting partner 3 |
0.172 |
0.054 |
| 48 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.612 |
-0.594 |
| 49 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.521 |
-0.049 |
| 50 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.490 |
-0.112 |
| 51 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.422 |
-0.056 |
| 52 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.363 |
-0.344 |
| 53 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.353 |
-0.753 |
| 54 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.347 |
-0.527 |
| 55 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.303 |
-0.128 |
| 56 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.301 |
-0.056 |
| 57 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.298 |
-0.199 |
| 58 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.296 |
-0.069 |
| 59 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.288 |
-0.098 |
| 60 |
247425_at |
AT5G62550
|
unknown |
-0.288 |
0.262 |
| 61 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.267 |
-0.573 |
| 62 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.262 |
-0.708 |
| 63 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.259 |
-0.366 |
| 64 |
265096_at |
AT1G04030
|
unknown |
-0.257 |
-0.091 |
| 65 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.243 |
0.167 |
| 66 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
-0.242 |
0.182 |
| 67 |
263282_at |
AT2G14095
|
unknown |
-0.236 |
-0.420 |
| 68 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.231 |
-0.519 |
| 69 |
258718_at |
AT3G09760
|
[RING/U-box superfamily protein] |
-0.228 |
-0.046 |
| 70 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.224 |
-0.801 |
| 71 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.216 |
-0.181 |
| 72 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.214 |
-0.216 |
| 73 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.208 |
0.059 |
| 74 |
252089_at |
AT3G52110
|
unknown |
-0.205 |
0.037 |
| 75 |
249060_at |
AT5G44560
|
VPS2.2 |
-0.204 |
0.482 |
| 76 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.197 |
0.004 |
| 77 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
-0.197 |
-0.261 |
| 78 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.197 |
-0.273 |
| 79 |
254670_at |
AT4G18390
|
TCP2, TCP2, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 |
-0.191 |
0.034 |
| 80 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.191 |
-0.215 |
| 81 |
248343_at |
AT5G52260
|
AtMYB19, myb domain protein 19, MYB19, myb domain protein 19 |
-0.186 |
-0.313 |
| 82 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
-0.184 |
-0.143 |
| 83 |
250369_at |
AT5G11300
|
CYC2BAT, CYC3B, mitotic-like cyclin 3B from Arabidopsis, CYCA2;2, CYCLIN A2;2 |
-0.180 |
0.012 |
| 84 |
255338_at |
AT4G04470
|
PMP22 |
-0.177 |
-0.005 |
| 85 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-0.175 |
-0.203 |
| 86 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.174 |
-0.432 |
| 87 |
260776_at |
AT1G14580
|
[C2H2-like zinc finger protein] |
-0.172 |
0.027 |
| 88 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.171 |
-0.078 |
| 89 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
-0.171 |
-0.062 |
| 90 |
245309_at |
AT4G15140
|
unknown |
-0.167 |
-0.093 |
| 91 |
265913_at |
AT2G25625
|
unknown |
-0.164 |
0.101 |
| 92 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.164 |
-0.012 |
| 93 |
261255_at |
AT1G05900
|
ATNTH2, NTH2, endonuclease III 2 |
-0.161 |
-0.012 |