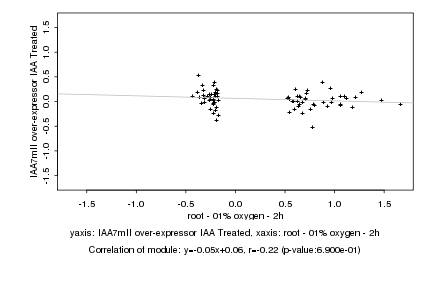
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root - 01% oxygen - 2h |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
250464_at |
AT5G10040
|
unknown |
1.661 |
-0.059 |
| 2 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.467 |
0.034 |
| 3 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
1.270 |
0.191 |
| 4 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
1.201 |
0.086 |
| 5 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
1.176 |
-0.114 |
| 6 |
254200_at |
AT4G24110
|
unknown |
1.113 |
0.078 |
| 7 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
1.100 |
0.113 |
| 8 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
1.058 |
-0.057 |
| 9 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
1.056 |
0.121 |
| 10 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
1.055 |
-0.076 |
| 11 |
249384_at |
AT5G39890
|
unknown |
0.969 |
0.078 |
| 12 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.965 |
-0.004 |
| 13 |
257925_at |
AT3G23170
|
unknown |
0.956 |
0.273 |
| 14 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.922 |
-0.092 |
| 15 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.886 |
-0.005 |
| 16 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.873 |
0.403 |
| 17 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.790 |
-0.065 |
| 18 |
250152_at |
AT5G15120
|
unknown |
0.784 |
-0.058 |
| 19 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.776 |
-0.515 |
| 20 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.754 |
-0.147 |
| 21 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.717 |
0.229 |
| 22 |
245226_at |
AT3G29970
|
[B12D protein] |
0.713 |
0.172 |
| 23 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.702 |
0.074 |
| 24 |
253616_at |
AT4G30380
|
[Barwin-related endoglucanase] |
0.700 |
0.042 |
| 25 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.672 |
-0.231 |
| 26 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.667 |
-0.005 |
| 27 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.649 |
0.084 |
| 28 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.638 |
0.113 |
| 29 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.638 |
-0.048 |
| 30 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
0.633 |
-0.099 |
| 31 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.619 |
0.004 |
| 32 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.616 |
0.108 |
| 33 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.601 |
0.253 |
| 34 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
0.588 |
-0.157 |
| 35 |
261567_at |
AT1G33055
|
unknown |
0.577 |
0.007 |
| 36 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.566 |
0.004 |
| 37 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.543 |
-0.214 |
| 38 |
266486_at |
AT2G47950
|
unknown |
0.541 |
0.053 |
| 39 |
263850_at |
AT2G04480
|
unknown |
0.527 |
0.085 |
| 40 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.522 |
0.075 |
| 41 |
254632_at |
AT4G18630
|
unknown |
-0.434 |
0.119 |
| 42 |
260883_at |
AT1G29270
|
unknown |
-0.383 |
0.186 |
| 43 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.377 |
0.536 |
| 44 |
252594_at |
AT3G45680
|
[Major facilitator superfamily protein] |
-0.367 |
0.100 |
| 45 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.343 |
-0.022 |
| 46 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.333 |
0.339 |
| 47 |
257056_at |
AT3G15350
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.325 |
0.138 |
| 48 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.325 |
0.232 |
| 49 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.320 |
0.075 |
| 50 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.315 |
-0.011 |
| 51 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.289 |
0.119 |
| 52 |
260067_at |
AT1G73780
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.268 |
0.147 |
| 53 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.265 |
0.037 |
| 54 |
262396_at |
AT1G49470
|
unknown |
-0.256 |
0.056 |
| 55 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.252 |
0.092 |
| 56 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.252 |
-0.153 |
| 57 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.246 |
0.161 |
| 58 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.230 |
-0.024 |
| 59 |
246534_at |
AT5G15890
|
TBL21, TRICHOME BIREFRINGENCE-LIKE 21 |
-0.229 |
-0.225 |
| 60 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.228 |
0.340 |
| 61 |
259220_at |
AT3G03550
|
[RING/U-box superfamily protein] |
-0.227 |
-0.053 |
| 62 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.225 |
0.047 |
| 63 |
245677_at |
AT1G56660
|
unknown |
-0.223 |
0.028 |
| 64 |
260024_at |
AT1G30080
|
[Glycosyl hydrolase superfamily protein] |
-0.218 |
-0.019 |
| 65 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
-0.218 |
-0.013 |
| 66 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.215 |
0.389 |
| 67 |
253195_at |
AT4G35420
|
DRL1, dihydroflavonol 4-reductase-like1, TKPR1, tetraketide alpha-pyrone reductase 1 |
-0.213 |
0.161 |
| 68 |
257670_at |
AT3G20340
|
unknown |
-0.212 |
0.161 |
| 69 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.210 |
0.115 |
| 70 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.208 |
-0.172 |
| 71 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.205 |
0.192 |
| 72 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.200 |
-0.111 |
| 73 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.200 |
-0.367 |
| 74 |
259520_at |
AT1G12320
|
unknown |
-0.198 |
0.249 |
| 75 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.188 |
0.165 |
| 76 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.182 |
0.229 |
| 77 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
-0.182 |
0.118 |
| 78 |
257311_at |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
-0.177 |
0.034 |
| 79 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
-0.176 |
-0.273 |