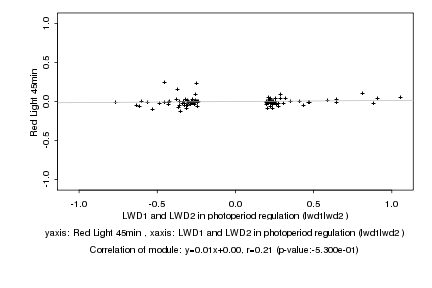
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
LWD1 and LWD2 in photoperiod regulation (lwd1lwd2 ) |
Log2 signal ratio
Red Light 45min |
| 1 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
1.053 |
0.057 |
| 2 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.908 |
0.045 |
| 3 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.882 |
-0.019 |
| 4 |
253322_at |
AT4G33980
|
unknown |
0.809 |
0.104 |
| 5 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.642 |
0.028 |
| 6 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.641 |
-0.004 |
| 7 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.588 |
0.014 |
| 8 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.470 |
-0.002 |
| 9 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.463 |
-0.001 |
| 10 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.435 |
-0.051 |
| 11 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
0.409 |
0.005 |
| 12 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.348 |
0.011 |
| 13 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.314 |
0.043 |
| 14 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
0.302 |
-0.019 |
| 15 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.287 |
0.091 |
| 16 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.286 |
0.039 |
| 17 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
0.273 |
-0.014 |
| 18 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.272 |
-0.062 |
| 19 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
0.260 |
-0.036 |
| 20 |
257076_at |
AT3G19680
|
unknown |
0.257 |
-0.034 |
| 21 |
252899_at |
AT4G39530
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.254 |
0.008 |
| 22 |
258225_at |
AT3G15630
|
unknown |
0.250 |
0.045 |
| 23 |
247280_at |
AT5G64260
|
EXL2, EXORDIUM like 2 |
0.248 |
-0.024 |
| 24 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.240 |
0.012 |
| 25 |
262211_at |
AT1G74930
|
ORA47 |
0.238 |
-0.014 |
| 26 |
259377_at |
AT3G16380
|
PAB6, poly(A) binding protein 6 |
0.238 |
0.006 |
| 27 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.234 |
-0.027 |
| 28 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
0.234 |
0.000 |
| 29 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.233 |
-0.035 |
| 30 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.233 |
-0.082 |
| 31 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.233 |
0.003 |
| 32 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
0.227 |
-0.007 |
| 33 |
245084_at |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
0.226 |
0.011 |
| 34 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
0.220 |
-0.002 |
| 35 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
0.219 |
0.051 |
| 36 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.219 |
-0.055 |
| 37 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.215 |
0.006 |
| 38 |
265900_at |
AT2G25730
|
unknown |
0.214 |
0.011 |
| 39 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.212 |
0.037 |
| 40 |
246212_at |
AT4G36930
|
SPT, SPATULA |
0.212 |
0.023 |
| 41 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.211 |
0.000 |
| 42 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
0.211 |
0.056 |
| 43 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.211 |
-0.019 |
| 44 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
0.206 |
-0.020 |
| 45 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.205 |
-0.082 |
| 46 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.203 |
-0.020 |
| 47 |
262503_at |
AT1G21670
|
[LOCATED IN: cell wall, plant-type cell wall] |
0.201 |
-0.015 |
| 48 |
251735_at |
AT3G56090
|
ATFER3, ferritin 3, FER3, ferritin 3 |
0.197 |
-0.029 |
| 49 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.196 |
-0.011 |
| 50 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.196 |
0.010 |
| 51 |
261202_at |
AT1G12910
|
ATAN11, ANTHOCYANIN11, LWD1, LIGHT-REGULATED WD 1 |
-0.770 |
-0.008 |
| 52 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.635 |
-0.040 |
| 53 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.615 |
-0.058 |
| 54 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.603 |
0.005 |
| 55 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.565 |
-0.011 |
| 56 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.536 |
-0.094 |
| 57 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.489 |
-0.018 |
| 58 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.454 |
-0.007 |
| 59 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.454 |
0.248 |
| 60 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.431 |
-0.006 |
| 61 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.429 |
-0.027 |
| 62 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.422 |
0.004 |
| 63 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.379 |
0.030 |
| 64 |
256926_at |
AT3G22540
|
unknown |
-0.371 |
0.162 |
| 65 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.365 |
-0.064 |
| 66 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.364 |
-0.040 |
| 67 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.363 |
0.011 |
| 68 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.356 |
-0.116 |
| 69 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.339 |
-0.015 |
| 70 |
257710_at |
AT3G27350
|
unknown |
-0.334 |
0.013 |
| 71 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.330 |
-0.024 |
| 72 |
261456_at |
AT1G21050
|
unknown |
-0.326 |
-0.040 |
| 73 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.323 |
0.033 |
| 74 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
-0.315 |
-0.085 |
| 75 |
255627_at |
AT4G00955
|
unknown |
-0.314 |
-0.039 |
| 76 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.312 |
0.019 |
| 77 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.310 |
-0.039 |
| 78 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.309 |
-0.046 |
| 79 |
252917_at |
AT4G38960
|
BBX19, B-box domain protein 19 |
-0.304 |
0.010 |
| 80 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
-0.304 |
-0.011 |
| 81 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.290 |
-0.029 |
| 82 |
249134_at |
AT5G43150
|
unknown |
-0.286 |
0.000 |
| 83 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.283 |
0.008 |
| 84 |
250744_at |
AT5G05840
|
unknown |
-0.282 |
-0.016 |
| 85 |
247686_at |
AT5G59700
|
[Protein kinase superfamily protein] |
-0.277 |
0.027 |
| 86 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.276 |
-0.023 |
| 87 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
-0.274 |
-0.019 |
| 88 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.274 |
-0.018 |
| 89 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.265 |
-0.033 |
| 90 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.265 |
0.001 |
| 91 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
-0.261 |
0.098 |
| 92 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.259 |
0.010 |
| 93 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.258 |
0.018 |
| 94 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.250 |
0.232 |
| 95 |
256674_at |
AT3G52360
|
unknown |
-0.250 |
0.005 |
| 96 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.245 |
-0.009 |
| 97 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.244 |
-0.053 |
| 98 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
-0.244 |
0.006 |
| 99 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.241 |
0.004 |