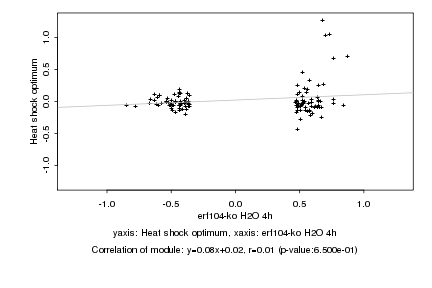
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
erf104-ko H2O 4h |
Log2 signal ratio
Heat shock optimum |
| 1 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.868 |
0.711 |
| 2 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.835 |
-0.053 |
| 3 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.762 |
0.680 |
| 4 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.758 |
-0.023 |
| 5 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.757 |
0.037 |
| 6 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.731 |
1.048 |
| 7 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.696 |
1.047 |
| 8 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.679 |
0.268 |
| 9 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.676 |
1.282 |
| 10 |
251699_at |
AT3G56630
|
CYP94D2, cytochrome P450, family 94, subfamily D, polypeptide 2 |
0.668 |
-0.082 |
| 11 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.667 |
-0.235 |
| 12 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.658 |
0.013 |
| 13 |
249271_at |
AT5G41790
|
CIP1, COP1-interactive protein 1 |
0.652 |
-0.092 |
| 14 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.646 |
0.004 |
| 15 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.642 |
0.017 |
| 16 |
257798_at |
AT3G15950
|
NAI2 |
0.642 |
0.266 |
| 17 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.639 |
-0.047 |
| 18 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.636 |
-0.080 |
| 19 |
262236_at |
AT1G48330
|
unknown |
0.635 |
0.076 |
| 20 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.621 |
-0.069 |
| 21 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.611 |
-0.084 |
| 22 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.595 |
-0.172 |
| 23 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
0.591 |
-0.012 |
| 24 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
0.590 |
-0.065 |
| 25 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.587 |
-0.002 |
| 26 |
250975_at |
AT5G03050
|
unknown |
0.586 |
0.045 |
| 27 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.583 |
-0.207 |
| 28 |
256096_at |
AT1G13650
|
unknown |
0.575 |
-0.151 |
| 29 |
260076_at |
AT1G73630
|
[EF hand calcium-binding protein family] |
0.574 |
-0.134 |
| 30 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.574 |
0.336 |
| 31 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.562 |
-0.030 |
| 32 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.561 |
0.193 |
| 33 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.558 |
-0.152 |
| 34 |
264958_at |
AT1G76960
|
unknown |
0.551 |
0.153 |
| 35 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
0.544 |
-0.089 |
| 36 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.538 |
-0.135 |
| 37 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.530 |
0.209 |
| 38 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
0.530 |
0.002 |
| 39 |
262693_at |
AT1G62780
|
unknown |
0.528 |
-0.019 |
| 40 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.524 |
-0.014 |
| 41 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.518 |
0.091 |
| 42 |
266903_at |
AT2G34570
|
MEE21, maternal effect embryo arrest 21 |
0.518 |
0.026 |
| 43 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.518 |
0.457 |
| 44 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.514 |
-0.048 |
| 45 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.512 |
-0.044 |
| 46 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.506 |
-0.281 |
| 47 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.505 |
-0.066 |
| 48 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.502 |
-0.126 |
| 49 |
256383_at |
AT1G66820
|
[glycine-rich protein] |
0.499 |
-0.065 |
| 50 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.496 |
0.149 |
| 51 |
257925_at |
AT3G23170
|
unknown |
0.489 |
-0.066 |
| 52 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
0.487 |
-0.026 |
| 53 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.484 |
-0.026 |
| 54 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.483 |
-0.437 |
| 55 |
258566_at |
AT3G04110
|
ATGLR1.1, GLR1, GLUTAMATE RECEPTOR 1, GLR1.1, glutamate receptor 1.1 |
0.482 |
-0.086 |
| 56 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.479 |
0.122 |
| 57 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.478 |
-0.126 |
| 58 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
0.477 |
-0.050 |
| 59 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
0.477 |
0.264 |
| 60 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.473 |
0.023 |
| 61 |
245937_at |
AT5G19750
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.470 |
0.003 |
| 62 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.469 |
-0.055 |
| 63 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.469 |
-0.159 |
| 64 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.464 |
-0.008 |
| 65 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
-0.850 |
-0.055 |
| 66 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
-0.783 |
-0.063 |
| 67 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.672 |
-0.022 |
| 68 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.666 |
0.046 |
| 69 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.634 |
0.026 |
| 70 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.632 |
0.114 |
| 71 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.615 |
-0.036 |
| 72 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.612 |
0.077 |
| 73 |
250624_at |
AT5G07330
|
unknown |
-0.600 |
-0.050 |
| 74 |
254263_at |
AT4G23493
|
unknown |
-0.595 |
0.106 |
| 75 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.592 |
0.103 |
| 76 |
255050_at |
AT4G09700
|
unknown |
-0.579 |
-0.017 |
| 77 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.543 |
0.004 |
| 78 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.541 |
0.012 |
| 79 |
254336_at |
AT4G22050
|
[Eukaryotic aspartyl protease family protein] |
-0.538 |
-0.005 |
| 80 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
-0.529 |
0.054 |
| 81 |
254635_at |
AT4G18670
|
[Leucine-rich repeat (LRR) family protein] |
-0.524 |
0.000 |
| 82 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
-0.517 |
-0.059 |
| 83 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.508 |
-0.031 |
| 84 |
256632_at |
AT3G28330
|
[F-box family protein-related] |
-0.507 |
-0.047 |
| 85 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
-0.504 |
-0.034 |
| 86 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
-0.503 |
0.025 |
| 87 |
245353_at |
AT4G16000
|
unknown |
-0.500 |
-0.109 |
| 88 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.496 |
-0.061 |
| 89 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.495 |
-0.129 |
| 90 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.485 |
-0.053 |
| 91 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.482 |
0.123 |
| 92 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.473 |
0.011 |
| 93 |
246693_at |
AT5G29070
|
unknown |
-0.472 |
0.003 |
| 94 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
-0.470 |
-0.170 |
| 95 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.467 |
-0.004 |
| 96 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.451 |
0.089 |
| 97 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
-0.443 |
-0.100 |
| 98 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.442 |
0.145 |
| 99 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.441 |
-0.055 |
| 100 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.440 |
0.138 |
| 101 |
259235_at |
AT3G11600
|
unknown |
-0.438 |
0.193 |
| 102 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-0.436 |
-0.050 |
| 103 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.436 |
-0.131 |
| 104 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
-0.433 |
-0.024 |
| 105 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.433 |
-0.033 |
| 106 |
250373_at |
AT5G11470
|
ASI1, ANTI-SILENCING 1, IBM2, Increase in Bonsai Methylation 2 |
-0.432 |
-0.052 |
| 107 |
267254_at |
AT2G23030
|
SNRK2-9, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-9, SNRK2.9, SNF1-related protein kinase 2.9 |
-0.430 |
0.009 |
| 108 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.430 |
-0.018 |
| 109 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.429 |
0.132 |
| 110 |
262764_at |
AT1G13140
|
CYP86C3, cytochrome P450, family 86, subfamily C, polypeptide 3 |
-0.420 |
-0.030 |
| 111 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
-0.413 |
-0.117 |
| 112 |
252675_at |
AT3G44270
|
[copia-like retrotransposon family, has a 1.1e-26 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.399 |
-0.020 |
| 113 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.398 |
0.028 |
| 114 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.394 |
-0.007 |
| 115 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.391 |
-0.189 |
| 116 |
245648_at |
AT1G24938
|
unknown |
-0.389 |
-0.065 |
| 117 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.388 |
0.048 |
| 118 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.385 |
-0.119 |
| 119 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.378 |
0.130 |
| 120 |
264311_at |
AT1G70400
|
unknown |
-0.370 |
-0.038 |
| 121 |
259984_at |
AT1G76460
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.370 |
0.014 |
| 122 |
259994_at |
AT1G68130
|
AtIDD14, indeterminate(ID)-domain 14, IDD14, indeterminate(ID)-domain 14, IDD14alpha, IDD14beta |
-0.369 |
-0.077 |
| 123 |
258745_at |
AT3G05920
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.367 |
0.009 |
| 124 |
255741_at |
AT1G25410
|
ATIPT6, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 6, IPT6, isopentenyltransferase 6 |
-0.367 |
-0.023 |
| 125 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.365 |
-0.034 |
| 126 |
259963_at |
AT1G53660
|
[nodulin MtN21 /EamA-like transporter family protein] |
-0.362 |
-0.071 |
| 127 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.360 |
0.102 |