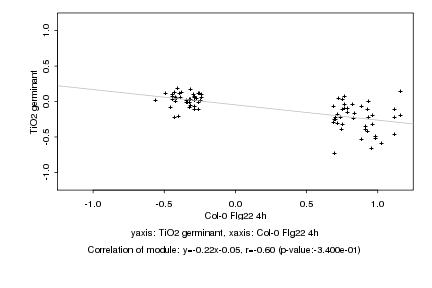
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-0 Flg22 4h |
Log2 signal ratio
TiO2 germinant |
| 1 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
1.157 |
0.145 |
| 2 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
1.154 |
-0.197 |
| 3 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
1.117 |
-0.219 |
| 4 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
1.116 |
-0.111 |
| 5 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.115 |
-0.457 |
| 6 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.022 |
-0.579 |
| 7 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.981 |
-0.481 |
| 8 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.979 |
-0.516 |
| 9 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.961 |
-0.322 |
| 10 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.959 |
-0.190 |
| 11 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.953 |
-0.660 |
| 12 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.932 |
0.013 |
| 13 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.929 |
-0.225 |
| 14 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.927 |
-0.108 |
| 15 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.926 |
-0.414 |
| 16 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.911 |
-0.340 |
| 17 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.911 |
-0.390 |
| 18 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.884 |
-0.062 |
| 19 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.883 |
-0.533 |
| 20 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.832 |
-0.161 |
| 21 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.829 |
-0.233 |
| 22 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.822 |
-0.039 |
| 23 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.785 |
-0.096 |
| 24 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
0.781 |
-0.150 |
| 25 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.766 |
0.080 |
| 26 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.765 |
-0.037 |
| 27 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.765 |
-0.093 |
| 28 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.750 |
-0.313 |
| 29 |
254408_at |
AT4G21390
|
B120 |
0.750 |
0.038 |
| 30 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.748 |
-0.101 |
| 31 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.743 |
-0.388 |
| 32 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.735 |
-0.221 |
| 33 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.723 |
0.046 |
| 34 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.716 |
-0.177 |
| 35 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.716 |
-0.301 |
| 36 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.702 |
-0.247 |
| 37 |
245209_at |
AT5G12340
|
unknown |
0.701 |
-0.222 |
| 38 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.696 |
-0.732 |
| 39 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.695 |
-0.245 |
| 40 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.688 |
-0.285 |
| 41 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.684 |
-0.062 |
| 42 |
254943_at |
AT4G10890
|
unknown |
-0.568 |
0.017 |
| 43 |
250565_at |
AT5G08000
|
E13L3, glucan endo-1,3-beta-glucosidase-like protein 3, PDCB2, PLASMODESMATA CALLOSE-BINDING PROTEIN 2 |
-0.493 |
0.123 |
| 44 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.460 |
-0.077 |
| 45 |
250156_at |
AT5G15170
|
TDP1, tyrosyl-DNA phosphodiesterase 1 |
-0.449 |
0.040 |
| 46 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.446 |
0.100 |
| 47 |
265131_at |
AT1G23760
|
JP630, PG3, POLYGALACTURONASE 3 |
-0.444 |
0.073 |
| 48 |
250882_at |
AT5G04000
|
unknown |
-0.430 |
-0.219 |
| 49 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.430 |
0.136 |
| 50 |
245648_at |
AT1G24938
|
unknown |
-0.425 |
0.013 |
| 51 |
267560_at |
AT2G45580
|
CYP76C3, cytochrome P450, family 76, subfamily C, polypeptide 3 |
-0.425 |
0.064 |
| 52 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
-0.416 |
0.054 |
| 53 |
247800_at |
AT5G58570
|
unknown |
-0.412 |
0.187 |
| 54 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
-0.406 |
-0.200 |
| 55 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.399 |
0.117 |
| 56 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.388 |
0.067 |
| 57 |
263674_at |
AT2G04790
|
unknown |
-0.380 |
0.135 |
| 58 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
-0.348 |
-0.013 |
| 59 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
-0.344 |
0.028 |
| 60 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.339 |
-0.012 |
| 61 |
247043_at |
AT5G66880
|
SNRK2-3, SUCROSE NONFERMENTING 1 (SNF1)-RELATED PROTEIN KINASE 2-3, SNRK2.3, sucrose nonfermenting 1(SNF1)-related protein kinase 2.3, SRK2I |
-0.329 |
0.038 |
| 62 |
247938_at |
AT5G57140
|
ATPAP28, PURPLE ACID PHOSPHATASE 28, PAP28, purple acid phosphatase 28 |
-0.325 |
-0.083 |
| 63 |
256006_at |
AT1G34070
|
unknown |
-0.324 |
-0.003 |
| 64 |
257517_at |
AT3G16330
|
unknown |
-0.319 |
-0.056 |
| 65 |
253141_at |
AT4G35440
|
ATCLC-E, CLC-E, chloride channel E, CLCE, CHLORIDE CHANNEL E |
-0.319 |
0.171 |
| 66 |
259698_at |
AT1G68930
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.318 |
0.034 |
| 67 |
247179_at |
AT5G65320
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.297 |
0.102 |
| 68 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.293 |
-0.112 |
| 69 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.292 |
-0.057 |
| 70 |
264311_at |
AT1G70400
|
unknown |
-0.292 |
0.062 |
| 71 |
259696_at |
AT1G63150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.291 |
0.026 |
| 72 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.288 |
0.082 |
| 73 |
253563_at |
AT4G31150
|
[endonuclease V family protein] |
-0.288 |
0.022 |
| 74 |
252761_at |
AT3G42770
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.276 |
0.050 |
| 75 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
-0.266 |
0.115 |
| 76 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.265 |
-0.013 |
| 77 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.262 |
-0.104 |
| 78 |
258573_at |
AT3G04260
|
PDE324, PIGMENT DEFECTIVE 324, PTAC3, plastid transcriptionally active 3 |
-0.256 |
0.116 |
| 79 |
252730_at |
AT3G43110
|
unknown |
-0.247 |
0.025 |
| 80 |
250285_at |
AT5G13300
|
AGD3, ARF-GAP DOMAIN3, SFC, SCARFACE, VAN3, ASCULAR NETWORK DEFECTIVE 3 |
-0.242 |
0.104 |
| 81 |
253028_at |
AT4G38160
|
pde191, pigment defective 191 |
-0.242 |
0.070 |