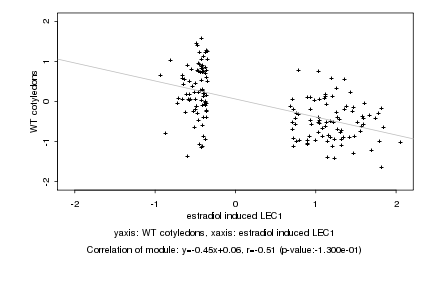
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
estradiol induced LEC1 |
Log2 signal ratio
WT cotyledons |
| 1 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
2.050 |
-1.013 |
| 2 |
265095_at |
AT1G03880
|
CRB, CRUCIFERIN B, CRU2, cruciferin 2 |
1.838 |
-0.647 |
| 3 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
1.816 |
-1.632 |
| 4 |
265094_at |
AT1G03890
|
[RmlC-like cupins superfamily protein] |
1.810 |
-0.165 |
| 5 |
253895_at |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
1.783 |
-0.980 |
| 6 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
1.772 |
-0.275 |
| 7 |
258327_at |
AT3G22640
|
PAP85 |
1.738 |
-0.401 |
| 8 |
251202_at |
AT3G63040
|
unknown |
1.684 |
-1.200 |
| 9 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
1.663 |
-0.347 |
| 10 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
1.601 |
-0.036 |
| 11 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
1.589 |
-0.555 |
| 12 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
1.587 |
-0.408 |
| 13 |
253241_at |
AT4G34520
|
FAE1, FATTY ACID ELONGATION1, KCS18, 3-ketoacyl-CoA synthase 18 |
1.571 |
-0.374 |
| 14 |
256290_at |
AT3G12203
|
scpl17, serine carboxypeptidase-like 17 |
1.569 |
-0.617 |
| 15 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
1.546 |
-0.747 |
| 16 |
262431_at |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
1.514 |
-0.511 |
| 17 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
1.475 |
-0.857 |
| 18 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
1.467 |
-1.281 |
| 19 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
1.465 |
-0.131 |
| 20 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
1.446 |
-0.233 |
| 21 |
260854_at |
AT1G21970
|
AtLEC1, EMB 212, EMBRYO DEFECTIVE 212, EMB212, EMBRYO DEFECTIVE 212, LEC1, LEAFY COTYLEDON 1, NF-YB9, NUCLEAR FACTOR Y, SUBUNIT B9 |
1.422 |
0.243 |
| 22 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
1.412 |
-0.878 |
| 23 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
1.372 |
-0.119 |
| 24 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
1.353 |
-0.191 |
| 25 |
255471_at |
AT4G03050
|
AOP3 |
1.353 |
0.568 |
| 26 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
1.337 |
-0.893 |
| 27 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
1.318 |
-0.948 |
| 28 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
1.312 |
-0.719 |
| 29 |
261915_at |
AT1G65880
|
BZO1, benzoyloxyglucosinolate 1 |
1.309 |
-1.095 |
| 30 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
1.300 |
-0.771 |
| 31 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
1.285 |
-0.441 |
| 32 |
263123_at |
AT1G78500
|
PEN6, PENTACYCLIC TRITERPENE SYNTHASE 6 |
1.259 |
-0.681 |
| 33 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
1.259 |
-0.389 |
| 34 |
257869_at |
AT3G25160
|
[ER lumen protein retaining receptor family protein] |
1.255 |
0.338 |
| 35 |
249547_at |
AT5G38160
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.248 |
-0.264 |
| 36 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
1.226 |
-0.940 |
| 37 |
254036_at |
AT4G25980
|
[Peroxidase superfamily protein] |
1.223 |
-1.412 |
| 38 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
1.210 |
-0.516 |
| 39 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
1.202 |
-1.114 |
| 40 |
249043_at |
AT5G44360
|
[FAD-binding Berberine family protein] |
1.198 |
0.141 |
| 41 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
1.185 |
0.600 |
| 42 |
248647_at |
AT5G49190
|
ATSUS2, SSA, SUCROSE SYNTHASE FROM ARABIDOPSIS, SUS2, sucrose synthase 2 |
1.180 |
-0.480 |
| 43 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
1.179 |
-0.882 |
| 44 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
1.150 |
-0.832 |
| 45 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
1.145 |
-1.375 |
| 46 |
250583_at |
AT5G07500
|
AtTZF6, PEI1, TZF6, Tandem CCCH Zinc Finger protein 6 |
1.139 |
-0.985 |
| 47 |
263138_at |
AT1G65090
|
unknown |
1.128 |
-0.070 |
| 48 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
1.123 |
-0.522 |
| 49 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.121 |
0.193 |
| 50 |
248096_at |
AT5G55240
|
ATPXG2, ARABIDOPSIS THALIANA PEROXYGENASE 2 |
1.111 |
-0.616 |
| 51 |
256827_at |
AT3G18570
|
[Oleosin family protein] |
1.110 |
0.140 |
| 52 |
259782_at |
AT1G29680
|
unknown |
1.100 |
0.076 |
| 53 |
262185_at |
AT1G77950
|
AGL67, AGAMOUS-like 67 |
1.075 |
-0.672 |
| 54 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
1.072 |
-0.856 |
| 55 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
1.045 |
-0.541 |
| 56 |
248754_at |
AT5G47670
|
L1L, LEC1-LIKE, NF-YB6, nuclear factor Y, subunit B6 |
1.045 |
0.072 |
| 57 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
1.039 |
-0.464 |
| 58 |
255840_at |
AT2G33520
|
unknown |
1.032 |
-0.752 |
| 59 |
259728_at |
AT1G60970
|
[SNARE-like superfamily protein] |
1.030 |
-0.382 |
| 60 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
1.029 |
0.768 |
| 61 |
249913_at |
AT5G22810
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.025 |
-0.503 |
| 62 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.985 |
-0.961 |
| 63 |
258258_at |
AT3G26790
|
FUS3, FUSCA 3 |
0.984 |
0.029 |
| 64 |
250893_at |
AT5G03795
|
[Exostosin family protein] |
0.937 |
-0.559 |
| 65 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.929 |
0.111 |
| 66 |
248494_at |
AT5G50770
|
AtHSD6, hydroxysteroid dehydrogenase 6, HSD6, hydroxysteroid dehydrogenase 6 |
0.927 |
-0.192 |
| 67 |
264606_at |
AT1G04660
|
[glycine-rich protein] |
0.925 |
-0.452 |
| 68 |
260265_at |
AT1G68510
|
LBD42, LOB domain-containing protein 42 |
0.913 |
-0.874 |
| 69 |
265863_at |
AT2G01770
|
ATVIT1, VIT1, vacuolar iron transporter 1 |
0.896 |
-0.964 |
| 70 |
246889_at |
AT5G25470
|
[AP2/B3-like transcriptional factor family protein] |
0.894 |
-1.064 |
| 71 |
261503_at |
AT1G71691
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.894 |
0.114 |
| 72 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.889 |
-1.028 |
| 73 |
250519_at |
AT5G08460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.790 |
-0.965 |
| 74 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
0.779 |
0.793 |
| 75 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.776 |
-0.302 |
| 76 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
0.755 |
-0.297 |
| 77 |
253375_at |
AT4G33280
|
[AP2/B3-like transcriptional factor family protein] |
0.755 |
-0.977 |
| 78 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
0.746 |
-0.416 |
| 79 |
265644_at |
AT2G27380
|
ATEPR1, extensin proline-rich 1, EPR1, extensin proline-rich 1 |
0.737 |
-0.574 |
| 80 |
246054_at |
AT5G08360
|
unknown |
0.720 |
-0.191 |
| 81 |
262734_at |
AT1G28640
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.713 |
-0.908 |
| 82 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
0.713 |
-1.100 |
| 83 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.704 |
-0.509 |
| 84 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.700 |
-0.687 |
| 85 |
255924_at |
AT1G22170
|
[Phosphoglycerate mutase family protein] |
0.698 |
0.067 |
| 86 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
0.678 |
-0.101 |
| 87 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.941 |
0.672 |
| 88 |
247070_at |
AT5G66815
|
unknown |
-0.881 |
-0.782 |
| 89 |
245165_at |
AT2G33180
|
unknown |
-0.810 |
1.046 |
| 90 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.733 |
-0.044 |
| 91 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.716 |
0.077 |
| 92 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.671 |
0.663 |
| 93 |
266813_at |
AT2G44920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.666 |
0.582 |
| 94 |
251466_at |
AT3G59340
|
unknown |
-0.660 |
0.066 |
| 95 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.659 |
0.432 |
| 96 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.646 |
0.559 |
| 97 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-0.627 |
-0.270 |
| 98 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.619 |
0.189 |
| 99 |
264343_at |
AT1G11850
|
unknown |
-0.608 |
0.914 |
| 100 |
249756_at |
AT5G24313
|
unknown |
-0.601 |
-1.372 |
| 101 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.586 |
0.062 |
| 102 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.582 |
0.193 |
| 103 |
252603_at |
AT3G45050
|
unknown |
-0.579 |
0.525 |
| 104 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.577 |
0.032 |
| 105 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
-0.567 |
0.056 |
| 106 |
250789_at |
AT5G05630
|
PUT3, POLYAMINE UPTAKE TRANSPORTER 3, RMV1, resistant to methyl viologen 1 |
-0.558 |
0.810 |
| 107 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.542 |
0.395 |
| 108 |
251660_at |
AT3G57160
|
unknown |
-0.530 |
-0.240 |
| 109 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-0.521 |
-0.638 |
| 110 |
264636_at |
AT1G65490
|
unknown |
-0.517 |
0.236 |
| 111 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.509 |
0.070 |
| 112 |
264613_at |
AT1G04640
|
LIP2, lipoyltransferase 2 |
-0.506 |
0.474 |
| 113 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
-0.498 |
-0.187 |
| 114 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
-0.497 |
-0.103 |
| 115 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.493 |
1.454 |
| 116 |
244997_at |
ATCG00170
|
RPOC2 |
-0.492 |
-0.104 |
| 117 |
262370_at |
AT1G73090
|
unknown |
-0.482 |
-0.284 |
| 118 |
248962_at |
AT5G45680
|
ATFKBP13, FK506 BINDING PROTEIN 13, FKBP13, FK506-binding protein 13 |
-0.476 |
1.408 |
| 119 |
247702_at |
AT5G59500
|
[protein C-terminal S-isoprenylcysteine carboxyl O-methyltransferases] |
-0.474 |
0.797 |
| 120 |
257647_at |
AT3G25805
|
unknown |
-0.471 |
0.770 |
| 121 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
-0.468 |
-0.465 |
| 122 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.468 |
0.967 |
| 123 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.462 |
0.769 |
| 124 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.460 |
0.232 |
| 125 |
256856_at |
AT3G15110
|
unknown |
-0.458 |
0.938 |
| 126 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.457 |
-1.065 |
| 127 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.453 |
1.243 |
| 128 |
251442_at |
AT3G59980
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.447 |
0.298 |
| 129 |
249162_at |
AT5G42765
|
unknown |
-0.444 |
0.733 |
| 130 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.433 |
-1.133 |
| 131 |
267586_at |
AT2G41950
|
unknown |
-0.433 |
1.060 |
| 132 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.432 |
0.547 |
| 133 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
-0.430 |
0.045 |
| 134 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.428 |
0.885 |
| 135 |
258989_at |
AT3G08920
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.424 |
0.842 |
| 136 |
255447_at |
AT4G02790
|
EMB3129, EMBRYO DEFECTIVE 3129 |
-0.424 |
1.580 |
| 137 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.422 |
-1.121 |
| 138 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.422 |
-0.079 |
| 139 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.421 |
-0.592 |
| 140 |
259093_at |
AT3G04860
|
unknown |
-0.421 |
0.726 |
| 141 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.420 |
0.319 |
| 142 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
-0.419 |
0.774 |
| 143 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
-0.418 |
0.295 |
| 144 |
246797_at |
AT5G26790
|
unknown |
-0.416 |
0.901 |
| 145 |
247329_at |
AT5G64150
|
[RNA methyltransferase family protein] |
-0.416 |
0.850 |
| 146 |
263471_at |
AT2G31890
|
ATRAP, RAP |
-0.410 |
1.132 |
| 147 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.402 |
0.146 |
| 148 |
258538_at |
AT3G06950
|
[Pseudouridine synthase family protein] |
-0.400 |
0.763 |
| 149 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
-0.400 |
-0.381 |
| 150 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
-0.399 |
0.218 |
| 151 |
257337_at |
ATMG00060
|
NAD5, NADH DEHYDROGENASE SUBUNIT 5, NAD5.3, NADH DEHYDROGENASE SUBUNIT 5.3, NAD5C, NADH dehydrogenase subunit 5C |
-0.398 |
-0.853 |
| 152 |
245930_at |
AT5G09240
|
[ssDNA-binding transcriptional regulator] |
-0.392 |
-0.061 |
| 153 |
260072_at |
AT1G73650
|
[FUNCTIONS IN: oxidoreductase activity, acting on the CH-CH group of donors] |
-0.387 |
0.219 |
| 154 |
261042_at |
AT1G01200
|
ATRAB-A3, ARABIDOPSIS RAB GTPASE HOMOLOG A3, ATRABA3, RAB GTPase homolog A3, RABA3, RAB GTPase homolog A3 |
-0.382 |
-0.001 |
| 155 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.382 |
-0.936 |
| 156 |
263760_at |
AT2G21280
|
ATSULA, GC1, GIANT CHLOROPLAST 1, SULA |
-0.380 |
1.235 |
| 157 |
262127_at |
AT1G52550
|
unknown |
-0.380 |
0.009 |
| 158 |
253816_at |
AT4G28210
|
EMB1923, embryo defective 1923 |
-0.378 |
-0.095 |
| 159 |
266867_at |
AT2G45770
|
CPFTSY, FRD4, FERRIC CHELATE REDUCTASE DEFECTIVE 4 |
-0.377 |
0.708 |
| 160 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.375 |
0.861 |
| 161 |
263831_at |
AT2G40300
|
ATFER4, ferritin 4, FER4, ferritin 4 |
-0.372 |
0.611 |
| 162 |
253814_at |
AT4G28290
|
unknown |
-0.370 |
0.164 |
| 163 |
250135_at |
AT5G15360
|
unknown |
-0.368 |
-0.397 |
| 164 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
-0.367 |
-0.201 |
| 165 |
249554_at |
AT5G38290
|
[Peptidyl-tRNA hydrolase family protein] |
-0.365 |
1.298 |
| 166 |
261608_at |
AT1G49650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.365 |
-0.233 |
| 167 |
248586_at |
AT5G49610
|
[F-box family protein] |
-0.364 |
0.795 |
| 168 |
250075_at |
AT5G17670
|
[alpha/beta-Hydrolases superfamily protein] |
-0.361 |
-0.031 |
| 169 |
264653_at |
AT1G08980
|
AMI1, amidase 1, ATAMI1, AMIDASE-LIKE PROTEIN 1, ATTOC64-I, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I, TOC64-I, TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I |
-0.359 |
1.268 |
| 170 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.358 |
1.058 |
| 171 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.352 |
0.514 |