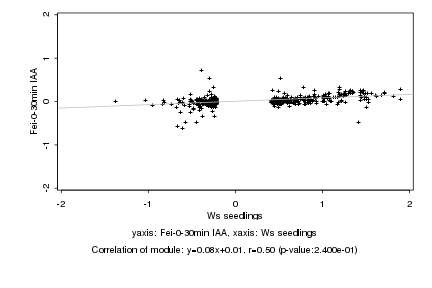
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ws seedlings |
Log2 signal ratio
Fei-0-30min IAA |
| 1 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.891 |
0.297 |
| 2 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.891 |
0.060 |
| 3 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
1.815 |
0.135 |
| 4 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
1.704 |
0.218 |
| 5 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
1.704 |
0.185 |
| 6 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
1.672 |
0.146 |
| 7 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
1.615 |
0.124 |
| 8 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
1.607 |
0.148 |
| 9 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
1.559 |
0.189 |
| 10 |
253895_at |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
1.519 |
-0.015 |
| 11 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.518 |
0.065 |
| 12 |
258327_at |
AT3G22640
|
PAP85 |
1.516 |
0.199 |
| 13 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
1.499 |
-0.136 |
| 14 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
1.496 |
0.107 |
| 15 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
1.494 |
0.056 |
| 16 |
247061_at |
AT5G66780
|
unknown |
1.488 |
0.194 |
| 17 |
265891_at |
AT2G15010
|
[Plant thionin] |
1.481 |
0.129 |
| 18 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
1.473 |
0.053 |
| 19 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
1.467 |
0.208 |
| 20 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.462 |
0.270 |
| 21 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
1.452 |
0.249 |
| 22 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
1.441 |
0.115 |
| 23 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
1.436 |
0.226 |
| 24 |
265095_at |
AT1G03880
|
CRB, CRUCIFERIN B, CRU2, cruciferin 2 |
1.435 |
0.039 |
| 25 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
1.433 |
0.210 |
| 26 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
1.433 |
0.260 |
| 27 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.429 |
0.149 |
| 28 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.402 |
-0.462 |
| 29 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
1.348 |
0.231 |
| 30 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
1.341 |
0.242 |
| 31 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
1.339 |
0.253 |
| 32 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
1.336 |
0.196 |
| 33 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
1.313 |
0.272 |
| 34 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
1.313 |
0.207 |
| 35 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
1.301 |
0.240 |
| 36 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
1.289 |
0.206 |
| 37 |
264494_at |
AT1G27461
|
unknown |
1.265 |
0.160 |
| 38 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
1.263 |
0.143 |
| 39 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
1.263 |
0.201 |
| 40 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
1.261 |
-0.006 |
| 41 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.256 |
0.254 |
| 42 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
1.252 |
0.193 |
| 43 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
1.235 |
0.121 |
| 44 |
249039_at |
AT5G44310
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.230 |
0.179 |
| 45 |
248915_at |
AT5G45690
|
unknown |
1.213 |
0.180 |
| 46 |
255049_at |
AT4G09610
|
GASA2, GAST1 protein homolog 2 |
1.210 |
0.021 |
| 47 |
263138_at |
AT1G65090
|
unknown |
1.207 |
0.042 |
| 48 |
259578_at |
AT1G27990
|
unknown |
1.195 |
0.158 |
| 49 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
1.194 |
0.172 |
| 50 |
251731_at |
AT3G56350
|
[Iron/manganese superoxide dismutase family protein] |
1.192 |
0.331 |
| 51 |
262828_at |
AT1G14950
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.190 |
0.287 |
| 52 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
1.189 |
0.136 |
| 53 |
250624_at |
AT5G07330
|
unknown |
1.180 |
0.210 |
| 54 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
1.178 |
-0.020 |
| 55 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
1.165 |
0.106 |
| 56 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
1.165 |
-0.046 |
| 57 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
1.141 |
0.104 |
| 58 |
253870_at |
AT4G27530
|
unknown |
1.117 |
0.094 |
| 59 |
262857_at |
AT1G14930
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.099 |
0.001 |
| 60 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
1.084 |
0.015 |
| 61 |
266392_at |
AT2G41280
|
ATM10, M10 |
1.080 |
0.084 |
| 62 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
1.071 |
0.194 |
| 63 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
1.070 |
0.028 |
| 64 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
1.068 |
0.107 |
| 65 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.061 |
0.202 |
| 66 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
1.059 |
0.121 |
| 67 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
1.050 |
0.072 |
| 68 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
1.042 |
0.104 |
| 69 |
265094_at |
AT1G03890
|
[RmlC-like cupins superfamily protein] |
1.035 |
-0.052 |
| 70 |
262310_at |
AT1G70840
|
MLP31, MLP-like protein 31 |
1.032 |
0.167 |
| 71 |
267579_at |
AT2G42000
|
AtMT4a, Arabidopsis thaliana metallothionein 4a |
1.021 |
0.087 |
| 72 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
1.020 |
0.117 |
| 73 |
249246_at |
AT5G42290
|
[transcription activator-related] |
1.020 |
0.020 |
| 74 |
245161_at |
AT2G33070
|
ATNSP2, NITRILE-SPECIFIER PROTEIN 2, NSP2, nitrile specifier protein 2 |
1.011 |
0.002 |
| 75 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
1.011 |
0.153 |
| 76 |
253494_at |
AT4G31830
|
unknown |
1.008 |
0.112 |
| 77 |
248096_at |
AT5G55240
|
ATPXG2, ARABIDOPSIS THALIANA PEROXYGENASE 2 |
1.005 |
0.070 |
| 78 |
257994_at |
AT3G19920
|
unknown |
1.004 |
0.134 |
| 79 |
267321_at |
AT2G19320
|
unknown |
0.994 |
0.013 |
| 80 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.951 |
0.129 |
| 81 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.922 |
-0.029 |
| 82 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.920 |
0.002 |
| 83 |
255840_at |
AT2G33520
|
unknown |
0.916 |
0.096 |
| 84 |
260917_at |
AT1G02700
|
unknown |
0.913 |
0.074 |
| 85 |
263881_at |
AT2G21820
|
unknown |
0.907 |
0.262 |
| 86 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.898 |
0.169 |
| 87 |
264888_at |
AT1G23070
|
unknown |
0.895 |
0.042 |
| 88 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.885 |
0.134 |
| 89 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.875 |
0.010 |
| 90 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.867 |
0.052 |
| 91 |
259615_at |
AT1G47980
|
unknown |
0.858 |
0.057 |
| 92 |
257059_at |
AT3G15280
|
unknown |
0.855 |
-0.035 |
| 93 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.853 |
0.029 |
| 94 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
0.852 |
0.001 |
| 95 |
250780_at |
AT5G05290
|
ATEXP2, ATEXPA2, expansin A2, ATHEXP ALPHA 1.12, EXP2, EXPANSIN 2, EXPA2, expansin A2 |
0.845 |
0.009 |
| 96 |
267036_at |
AT2G38465
|
unknown |
0.845 |
0.118 |
| 97 |
262185_at |
AT1G77950
|
AGL67, AGAMOUS-like 67 |
0.835 |
0.029 |
| 98 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.830 |
-0.035 |
| 99 |
265644_at |
AT2G27380
|
ATEPR1, extensin proline-rich 1, EPR1, extensin proline-rich 1 |
0.828 |
0.110 |
| 100 |
249364_at |
AT5G40590
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.824 |
0.105 |
| 101 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.822 |
0.032 |
| 102 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
0.821 |
-0.017 |
| 103 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.800 |
0.096 |
| 104 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.798 |
-0.047 |
| 105 |
265334_at |
AT2G18370
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.797 |
-0.006 |
| 106 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.790 |
0.046 |
| 107 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.787 |
-0.051 |
| 108 |
250010_at |
AT5G18450
|
[Integrase-type DNA-binding superfamily protein] |
0.783 |
0.060 |
| 109 |
262527_at |
AT1G17010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.782 |
0.076 |
| 110 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.780 |
0.330 |
| 111 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
0.778 |
0.040 |
| 112 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.772 |
0.080 |
| 113 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
0.768 |
0.048 |
| 114 |
260458_at |
AT1G68250
|
unknown |
0.747 |
0.104 |
| 115 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.745 |
0.049 |
| 116 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.743 |
0.154 |
| 117 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.732 |
-0.029 |
| 118 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.719 |
0.014 |
| 119 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.716 |
-0.053 |
| 120 |
259782_at |
AT1G29680
|
unknown |
0.709 |
0.068 |
| 121 |
253322_at |
AT4G33980
|
unknown |
0.708 |
0.094 |
| 122 |
259353_at |
AT3G05190
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.706 |
0.048 |
| 123 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.704 |
0.064 |
| 124 |
256827_at |
AT3G18570
|
[Oleosin family protein] |
0.702 |
0.006 |
| 125 |
260994_at |
AT1G12130
|
[Flavin-binding monooxygenase family protein] |
0.701 |
0.045 |
| 126 |
255493_at |
AT4G02690
|
AtLFG3, LFG3, LIFEGUARD 3 |
0.691 |
0.014 |
| 127 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.688 |
0.044 |
| 128 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.687 |
0.070 |
| 129 |
261936_at |
AT1G22600
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.683 |
0.063 |
| 130 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.682 |
0.017 |
| 131 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.681 |
0.015 |
| 132 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.679 |
-0.068 |
| 133 |
260859_at |
AT1G43780
|
scpl44, serine carboxypeptidase-like 44 |
0.676 |
0.018 |
| 134 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.671 |
0.051 |
| 135 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.669 |
0.025 |
| 136 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.669 |
-0.006 |
| 137 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.666 |
-0.026 |
| 138 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.662 |
0.056 |
| 139 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.639 |
0.103 |
| 140 |
247172_at |
AT5G65550
|
[UDP-Glycosyltransferase superfamily protein] |
0.638 |
-0.003 |
| 141 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
0.632 |
-0.002 |
| 142 |
267286_at |
AT2G23640
|
RTNLB13, Reticulan like protein B13 |
0.631 |
0.063 |
| 143 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
0.625 |
-0.020 |
| 144 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.621 |
0.035 |
| 145 |
248076_at |
AT5G55750
|
[hydroxyproline-rich glycoprotein family protein] |
0.617 |
-0.010 |
| 146 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.614 |
-0.097 |
| 147 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.598 |
0.008 |
| 148 |
249111_at |
AT5G43770
|
[proline-rich family protein] |
0.594 |
0.092 |
| 149 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.593 |
0.053 |
| 150 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
0.590 |
-0.028 |
| 151 |
255763_at |
AT1G16730
|
unknown |
0.589 |
0.155 |
| 152 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.586 |
-0.006 |
| 153 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
0.586 |
0.062 |
| 154 |
251202_at |
AT3G63040
|
unknown |
0.583 |
0.073 |
| 155 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.582 |
-0.010 |
| 156 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.581 |
-0.007 |
| 157 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
0.580 |
0.032 |
| 158 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.579 |
-0.015 |
| 159 |
256898_at |
AT3G24650
|
ABI3, ABA INSENSITIVE 3, ABI3, ABSCISIC ACID INSENSITIVE 3, SIS10, SUGAR INSENSITIVE 10 |
0.570 |
0.072 |
| 160 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.569 |
0.071 |
| 161 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.563 |
0.007 |
| 162 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.563 |
0.192 |
| 163 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.549 |
0.089 |
| 164 |
247269_at |
AT5G64210
|
AOX2, alternative oxidase 2 |
0.548 |
0.046 |
| 165 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.547 |
-0.001 |
| 166 |
252914_at |
AT4G39130
|
[Dehydrin family protein] |
0.546 |
-0.003 |
| 167 |
262069_at |
AT1G80090
|
CBSX4, CBS domain containing protein 4 |
0.546 |
0.051 |
| 168 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.545 |
0.019 |
| 169 |
263034_at |
AT1G24020
|
MLP423, MLP-like protein 423 |
0.542 |
0.023 |
| 170 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.541 |
0.113 |
| 171 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
0.536 |
0.027 |
| 172 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.534 |
0.007 |
| 173 |
257234_at |
AT3G14880
|
unknown |
0.533 |
0.077 |
| 174 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.532 |
-0.043 |
| 175 |
265714_at |
AT2G03520
|
ATUPS4, ureide permease 4, UPS4, ureide permease 4 |
0.530 |
0.029 |
| 176 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.530 |
0.003 |
| 177 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.529 |
0.057 |
| 178 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.524 |
-0.062 |
| 179 |
256114_at |
AT1G16850
|
unknown |
0.517 |
-0.053 |
| 180 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
0.514 |
0.001 |
| 181 |
245076_at |
AT2G23170
|
GH3.3 |
0.514 |
0.541 |
| 182 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.511 |
0.010 |
| 183 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.509 |
0.005 |
| 184 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.509 |
0.048 |
| 185 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.507 |
0.033 |
| 186 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.506 |
-0.042 |
| 187 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.504 |
0.037 |
| 188 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.503 |
0.067 |
| 189 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.501 |
0.042 |
| 190 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
0.499 |
-0.056 |
| 191 |
247883_at |
AT5G57790
|
unknown |
0.497 |
0.018 |
| 192 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
0.493 |
0.015 |
| 193 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
0.492 |
0.254 |
| 194 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.490 |
0.013 |
| 195 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.489 |
-0.122 |
| 196 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.487 |
-0.044 |
| 197 |
247904_at |
AT5G57390
|
AIL5, AINTEGUMENTA-like 5, CHO1, CHOTTO 1, EMK, EMBRYOMAKER, PLT5, PLETHORA 5 |
0.487 |
0.039 |
| 198 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.486 |
0.080 |
| 199 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.484 |
0.027 |
| 200 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
0.480 |
-0.062 |
| 201 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.477 |
0.037 |
| 202 |
254854_at |
AT4G12130
|
[Glycine cleavage T-protein family] |
0.476 |
-0.023 |
| 203 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.472 |
-0.020 |
| 204 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.471 |
0.029 |
| 205 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
0.471 |
-0.015 |
| 206 |
265099_at |
AT1G03990
|
[Long-chain fatty alcohol dehydrogenase family protein] |
0.469 |
0.101 |
| 207 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.463 |
0.003 |
| 208 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.462 |
0.072 |
| 209 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.460 |
-0.020 |
| 210 |
250882_at |
AT5G04000
|
unknown |
0.455 |
0.038 |
| 211 |
263907_at |
AT2G36270
|
ABI5, ABA INSENSITIVE 5, AtABI5, GIA1, GROWTH-INSENSITIVITY TO ABA 1 |
0.447 |
0.016 |
| 212 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.447 |
0.081 |
| 213 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.445 |
0.101 |
| 214 |
247762_at |
AT5G59170
|
[Proline-rich extensin-like family protein] |
0.444 |
0.037 |
| 215 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.443 |
-0.050 |
| 216 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.441 |
0.019 |
| 217 |
252898_at |
AT4G39500
|
CYP96A11, cytochrome P450, family 96, subfamily A, polypeptide 11 |
0.441 |
0.030 |
| 218 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.441 |
-0.035 |
| 219 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.440 |
0.032 |
| 220 |
250828_at |
AT5G05250
|
unknown |
0.438 |
-0.101 |
| 221 |
250426_at |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
0.434 |
0.043 |
| 222 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.433 |
0.029 |
| 223 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
0.430 |
-0.068 |
| 224 |
247096_at |
AT5G66430
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.429 |
0.047 |
| 225 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.428 |
0.006 |
| 226 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.428 |
0.021 |
| 227 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.428 |
-0.006 |
| 228 |
245209_at |
AT5G12340
|
unknown |
0.418 |
0.276 |
| 229 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.418 |
-0.013 |
| 230 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.417 |
0.040 |
| 231 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.416 |
0.014 |
| 232 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.416 |
-0.033 |
| 233 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
0.414 |
-0.008 |
| 234 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.414 |
0.029 |
| 235 |
255443_at |
AT4G02700
|
SULTR3;2, sulfate transporter 3;2 |
0.413 |
-0.020 |
| 236 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.411 |
0.045 |
| 237 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.411 |
0.007 |
| 238 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.410 |
-0.014 |
| 239 |
245226_at |
AT3G29970
|
[B12D protein] |
-1.387 |
0.006 |
| 240 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-1.033 |
0.033 |
| 241 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
-0.958 |
-0.088 |
| 242 |
247024_at |
AT5G66985
|
unknown |
-0.839 |
-0.062 |
| 243 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.837 |
0.027 |
| 244 |
250464_at |
AT5G10040
|
unknown |
-0.816 |
-0.000 |
| 245 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.745 |
-0.051 |
| 246 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.684 |
-0.131 |
| 247 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.675 |
0.050 |
| 248 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.675 |
-0.558 |
| 249 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.657 |
-0.028 |
| 250 |
267475_at |
AT2G02730
|
unknown |
-0.648 |
0.024 |
| 251 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.640 |
-0.003 |
| 252 |
250472_at |
AT5G10210
|
unknown |
-0.640 |
-0.237 |
| 253 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.618 |
-0.617 |
| 254 |
263607_at |
AT2G16270
|
unknown |
-0.617 |
-0.012 |
| 255 |
252882_at |
AT4G39675
|
unknown |
-0.609 |
-0.026 |
| 256 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.607 |
0.078 |
| 257 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
-0.588 |
-0.084 |
| 258 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.581 |
-0.468 |
| 259 |
257367_at |
AT2G25780
|
unknown |
-0.535 |
-0.062 |
| 260 |
263545_at |
AT2G21560
|
unknown |
-0.532 |
-0.093 |
| 261 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.527 |
-0.240 |
| 262 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
-0.523 |
0.002 |
| 263 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.520 |
0.174 |
| 264 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.518 |
0.049 |
| 265 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.513 |
0.042 |
| 266 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.504 |
-0.002 |
| 267 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
-0.498 |
-0.009 |
| 268 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
-0.493 |
-0.012 |
| 269 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.489 |
-0.141 |
| 270 |
249384_at |
AT5G39890
|
unknown |
-0.486 |
-0.171 |
| 271 |
266021_at |
AT2G05910
|
unknown |
-0.485 |
-0.038 |
| 272 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
-0.456 |
0.025 |
| 273 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.452 |
-0.064 |
| 274 |
261567_at |
AT1G33055
|
unknown |
-0.448 |
-0.473 |
| 275 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.448 |
0.057 |
| 276 |
253859_at |
AT4G27657
|
unknown |
-0.443 |
0.061 |
| 277 |
246344_at |
AT3G56730
|
[Putative endonuclease or glycosyl hydrolase] |
-0.442 |
0.034 |
| 278 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.433 |
-0.040 |
| 279 |
267474_at |
AT2G02740
|
ATWHY3, A. THALIANA WHIRLY 3, PTAC11, PLASTID TRANSCRIPTIONALLY ACTIVE11, WHY3, WHIRLY 3 |
-0.432 |
-0.020 |
| 280 |
258362_at |
AT3G14280
|
unknown |
-0.427 |
-0.076 |
| 281 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.423 |
0.083 |
| 282 |
252834_at |
AT4G40070
|
[RING/U-box superfamily protein] |
-0.422 |
0.004 |
| 283 |
265892_at |
AT2G15020
|
unknown |
-0.420 |
-0.056 |
| 284 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.419 |
-0.189 |
| 285 |
264909_at |
AT2G17300
|
unknown |
-0.417 |
-0.136 |
| 286 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.415 |
0.024 |
| 287 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.415 |
0.007 |
| 288 |
250152_at |
AT5G15120
|
unknown |
-0.408 |
-0.102 |
| 289 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
-0.397 |
0.031 |
| 290 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.397 |
0.020 |
| 291 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.396 |
0.726 |
| 292 |
252400_at |
AT3G48020
|
unknown |
-0.392 |
0.049 |
| 293 |
246142_at |
AT5G19970
|
unknown |
-0.391 |
-0.151 |
| 294 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
-0.390 |
-0.017 |
| 295 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
-0.389 |
-0.047 |
| 296 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.388 |
0.040 |
| 297 |
254200_at |
AT4G24110
|
unknown |
-0.386 |
-0.327 |
| 298 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
-0.385 |
0.020 |
| 299 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.383 |
0.002 |
| 300 |
257670_at |
AT3G20340
|
unknown |
-0.370 |
-0.057 |
| 301 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.368 |
0.008 |
| 302 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
-0.367 |
0.027 |
| 303 |
255806_at |
AT4G10260
|
[pfkB-like carbohydrate kinase family protein] |
-0.367 |
0.019 |
| 304 |
257945_at |
AT3G21860
|
ASK10, SKP1-like 10, SK10, SKP1-like 10 |
-0.361 |
0.004 |
| 305 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.359 |
0.006 |
| 306 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.359 |
-0.015 |
| 307 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.356 |
-0.026 |
| 308 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.356 |
0.103 |
| 309 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.354 |
0.004 |
| 310 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
-0.352 |
0.009 |
| 311 |
249069_at |
AT5G44010
|
unknown |
-0.350 |
0.044 |
| 312 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.350 |
-0.060 |
| 313 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
-0.350 |
-0.022 |
| 314 |
263109_at |
AT1G65180
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.347 |
-0.053 |
| 315 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
-0.347 |
0.007 |
| 316 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.344 |
-0.005 |
| 317 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.343 |
-0.046 |
| 318 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
-0.338 |
-0.014 |
| 319 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.335 |
-0.076 |
| 320 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
-0.333 |
-0.055 |
| 321 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
-0.333 |
0.024 |
| 322 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.329 |
0.141 |
| 323 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.327 |
-0.043 |
| 324 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.326 |
-0.002 |
| 325 |
257076_at |
AT3G19680
|
unknown |
-0.321 |
-0.027 |
| 326 |
261350_at |
AT1G79770
|
unknown |
-0.321 |
-0.026 |
| 327 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.320 |
0.004 |
| 328 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.318 |
0.019 |
| 329 |
261139_at |
AT1G19700
|
BEL10, BEL1-like homeodomain 10, BLH10, BEL1-LIKE HOMEODOMAIN 10 |
-0.317 |
-0.054 |
| 330 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
-0.316 |
0.013 |
| 331 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.315 |
-0.094 |
| 332 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
-0.314 |
-0.019 |
| 333 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
-0.311 |
-0.068 |
| 334 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.308 |
-0.051 |
| 335 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.306 |
0.008 |
| 336 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.306 |
0.239 |
| 337 |
246860_at |
AT5G25840
|
unknown |
-0.303 |
-0.028 |
| 338 |
265938_at |
AT2G19620
|
NDL3, N-MYC downregulated-like 3 |
-0.303 |
0.020 |
| 339 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.302 |
0.008 |
| 340 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.302 |
-0.063 |
| 341 |
261491_at |
AT1G14350
|
AtMYB124, myb domain protein 124, FLP, FOUR LIPS, MYB124 |
-0.302 |
-0.016 |
| 342 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.301 |
0.534 |
| 343 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
-0.301 |
0.059 |
| 344 |
267485_at |
AT2G02820
|
AtMYB88, myb domain protein 88, MYB88, myb domain protein 88 |
-0.298 |
-0.011 |
| 345 |
254681_at |
AT4G18140
|
SSP4b, SCP1-like small phosphatase 4b |
-0.297 |
0.015 |
| 346 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
-0.297 |
0.029 |
| 347 |
245825_at |
AT1G57870
|
ATSK42, shaggy-like kinase 42, SK42, shaggy-like kinase 42 |
-0.296 |
-0.023 |
| 348 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.295 |
0.003 |
| 349 |
247165_at |
AT5G65790
|
ATMYB68, MYB DOMAIN PROTEIN 68, MYB68, myb domain protein 68 |
-0.293 |
-0.060 |
| 350 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
-0.291 |
0.016 |
| 351 |
250881_at |
AT5G04080
|
unknown |
-0.291 |
0.008 |
| 352 |
261263_at |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
-0.291 |
-0.012 |
| 353 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.290 |
-0.061 |
| 354 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.290 |
0.008 |
| 355 |
265057_at |
AT1G52140
|
unknown |
-0.289 |
-0.081 |
| 356 |
245610_at |
AT4G14380
|
unknown |
-0.287 |
0.027 |
| 357 |
255909_at |
AT1G18040
|
AT;CDCKD;3, CYCLIN-DEPENDENT KINASE D1;3, CAK2AT, CDKD1;3, cyclin-dependent kinase D1;3 |
-0.287 |
0.010 |
| 358 |
255858_at |
AT1G67030
|
ZFP6, zinc finger protein 6 |
-0.286 |
-0.115 |
| 359 |
251293_at |
AT3G61930
|
unknown |
-0.286 |
0.036 |
| 360 |
260717_at |
AT1G48120
|
[hydrolases] |
-0.286 |
-0.066 |
| 361 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.286 |
-0.040 |
| 362 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.286 |
-0.012 |
| 363 |
264840_at |
AT1G03440
|
[Leucine-rich repeat (LRR) family protein] |
-0.284 |
0.003 |
| 364 |
251478_at |
AT3G59690
|
IQD13, IQ-domain 13 |
-0.283 |
0.032 |
| 365 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.283 |
0.169 |
| 366 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.282 |
0.099 |
| 367 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.278 |
0.007 |
| 368 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
-0.277 |
0.041 |
| 369 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.277 |
0.002 |
| 370 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
-0.277 |
0.023 |
| 371 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
-0.277 |
-0.074 |
| 372 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.276 |
-0.025 |
| 373 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.274 |
-0.048 |
| 374 |
266222_at |
AT2G28780
|
unknown |
-0.273 |
0.016 |
| 375 |
262450_at |
AT1G11320
|
unknown |
-0.272 |
-0.030 |
| 376 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.268 |
-0.223 |
| 377 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
-0.266 |
0.008 |
| 378 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.266 |
-0.041 |
| 379 |
264814_at |
AT2G17900
|
ASHR1, ASH1-related 1, SDG37, SET domain group 37 |
-0.265 |
0.019 |
| 380 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
-0.265 |
-0.031 |
| 381 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
-0.264 |
-0.084 |
| 382 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.264 |
-0.009 |
| 383 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.263 |
-0.105 |
| 384 |
267580_at |
AT2G41990
|
unknown |
-0.262 |
0.081 |
| 385 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.262 |
-0.092 |
| 386 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.262 |
0.011 |
| 387 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.262 |
-0.132 |
| 388 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.261 |
0.032 |
| 389 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.260 |
0.053 |
| 390 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.259 |
-0.121 |
| 391 |
250265_at |
AT5G12900
|
unknown |
-0.258 |
0.011 |
| 392 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.258 |
0.084 |
| 393 |
246385_at |
AT1G77390
|
CYCA1, CYCLIN A1, CYCA1;2, CYCLIN A1;2, DYP, TAM, TARDY ASYNCHRONOUS MEIOSIS |
-0.258 |
0.001 |
| 394 |
253892_at |
AT4G27620
|
unknown |
-0.257 |
0.031 |
| 395 |
254749_at |
AT4G13130
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.257 |
0.049 |
| 396 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.256 |
0.009 |
| 397 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.252 |
-0.039 |
| 398 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.252 |
-0.033 |
| 399 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-0.252 |
0.336 |
| 400 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
-0.251 |
0.010 |
| 401 |
249464_at |
AT5G39710
|
EMB2745, EMBRYO DEFECTIVE 2745 |
-0.251 |
-0.015 |
| 402 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.251 |
-0.046 |
| 403 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.250 |
-0.014 |
| 404 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
-0.250 |
0.060 |
| 405 |
251939_at |
AT3G53440
|
[Homeodomain-like superfamily protein] |
-0.250 |
-0.009 |
| 406 |
263956_at |
AT2G35940
|
BLH1, BEL1-like homeodomain 1, EDA29, embryo sac development arrest 29 |
-0.250 |
0.015 |
| 407 |
251437_at |
AT3G59910
|
[Ankyrin repeat family protein] |
-0.250 |
0.004 |
| 408 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.249 |
-0.340 |
| 409 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
-0.248 |
-0.037 |
| 410 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.247 |
-0.109 |
| 411 |
254178_at |
AT4G23880
|
unknown |
-0.247 |
0.018 |
| 412 |
246843_at |
AT5G26740
|
unknown |
-0.246 |
-0.023 |
| 413 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.246 |
0.032 |
| 414 |
261345_at |
AT1G79760
|
DTA4, downstream target of AGL15-4 |
-0.245 |
0.132 |
| 415 |
253517_at |
AT4G31390
|
ABC1K1, ABC1-like kinase 1, ACDO1, ABC1-like kinase related to chlorophyll degradation and oxidative stress 1, AtACDO1, PGR6, PROTON GRADIENT REGULATION 6 |
-0.245 |
0.018 |
| 416 |
262388_at |
AT1G49320
|
unknown |
-0.245 |
0.016 |
| 417 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
-0.245 |
0.073 |
| 418 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.245 |
0.061 |
| 419 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.245 |
0.016 |
| 420 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
-0.243 |
0.029 |
| 421 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.243 |
-0.002 |
| 422 |
249201_at |
AT5G42370
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.241 |
-0.040 |
| 423 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.241 |
-0.108 |
| 424 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
-0.241 |
-0.042 |
| 425 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
-0.241 |
-0.083 |
| 426 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.241 |
-0.048 |
| 427 |
260668_at |
AT1G19530
|
unknown |
-0.241 |
0.077 |
| 428 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
-0.240 |
0.015 |
| 429 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.240 |
-0.067 |
| 430 |
249016_at |
AT5G44750
|
ATREV1, ARABIDOPSIS THALIANA HOMOLOG OF REVERSIONLESS 1, REV1 |
-0.240 |
-0.044 |
| 431 |
264238_at |
AT1G54740
|
unknown |
-0.240 |
-0.120 |
| 432 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.239 |
0.020 |
| 433 |
258196_at |
AT3G13980
|
unknown |
-0.239 |
0.115 |
| 434 |
253386_at |
AT4G33030
|
SQD1, sulfoquinovosyldiacylglycerol 1 |
-0.238 |
0.010 |
| 435 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.237 |
0.051 |
| 436 |
259596_at |
AT1G28130
|
GH3.17 |
-0.237 |
-0.051 |
| 437 |
263674_at |
AT2G04790
|
unknown |
-0.236 |
-0.020 |
| 438 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.236 |
0.054 |
| 439 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
-0.235 |
0.002 |
| 440 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
-0.234 |
-0.002 |
| 441 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
-0.234 |
0.011 |
| 442 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.234 |
-0.057 |
| 443 |
261522_at |
AT1G71710
|
[DNAse I-like superfamily protein] |
-0.233 |
-0.011 |
| 444 |
249905_at |
AT5G22720
|
[F-box/RNI-like superfamily protein] |
-0.233 |
-0.008 |
| 445 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
-0.233 |
0.002 |
| 446 |
267232_at |
AT2G44190
|
EDE1, ENDOSPERM DEFECTIVE 1, EMB3116, EMBRYO DEFECTIVE 3116, QWRF5, QWRF domain containing 5 |
-0.233 |
0.041 |
| 447 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.232 |
0.040 |
| 448 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.232 |
0.016 |
| 449 |
255221_at |
AT4G05150
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.231 |
-0.036 |
| 450 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.231 |
-0.038 |
| 451 |
267178_at |
AT2G37750
|
unknown |
-0.231 |
-0.063 |
| 452 |
247994_at |
AT5G56140
|
[RNA-binding KH domain-containing protein] |
-0.230 |
0.021 |
| 453 |
264836_at |
AT1G03610
|
unknown |
-0.230 |
-0.046 |
| 454 |
259226_at |
AT3G07700
|
[Protein kinase superfamily protein] |
-0.229 |
0.010 |
| 455 |
258122_at |
AT3G14570
|
ATGSL04, glucan synthase-like 4, atgsl4, gsl04, GSL04, glucan synthase-like 4 |
-0.229 |
-0.056 |
| 456 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.228 |
0.044 |
| 457 |
255385_at |
AT4G03610
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.228 |
0.018 |
| 458 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
-0.228 |
-0.034 |
| 459 |
256734_at |
AT3G29390
|
RIK, RS2-interacting KH protein |
-0.228 |
-0.011 |
| 460 |
261700_at |
AT1G32690
|
unknown |
-0.228 |
0.016 |
| 461 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.227 |
-0.013 |
| 462 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.227 |
0.089 |
| 463 |
262158_at |
AT1G52540
|
[Protein kinase superfamily protein] |
-0.226 |
-0.028 |
| 464 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.226 |
-0.027 |
| 465 |
260273_at |
AT1G80550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.226 |
0.028 |
| 466 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.226 |
0.006 |
| 467 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
-0.226 |
0.063 |
| 468 |
261674_at |
AT1G18270
|
[ketose-bisphosphate aldolase class-II family protein] |
-0.225 |
-0.020 |
| 469 |
258221_at |
AT3G29160
|
AKIN11, SNF1 kinase homolog 11, ATKIN11, SNF1 kinase homolog 11, KIN11, SNF1 kinase homolog 11, SNRK1.2, SNF1-RELATED PROTEIN KINASE 1.2 |
-0.224 |
0.003 |
| 470 |
257007_at |
AT3G14205
|
[Phosphoinositide phosphatase family protein] |
-0.224 |
-0.009 |
| 471 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.223 |
0.050 |
| 472 |
259485_at |
AT1G15700
|
ATPC2 |
-0.223 |
0.003 |
| 473 |
265323_at |
AT2G18260
|
ATSYP112, SYP112, syntaxin of plants 112 |
-0.223 |
0.075 |
| 474 |
262127_at |
AT1G52550
|
unknown |
-0.223 |
-0.016 |
| 475 |
258326_at |
AT3G22760
|
SOL1 |
-0.222 |
0.006 |