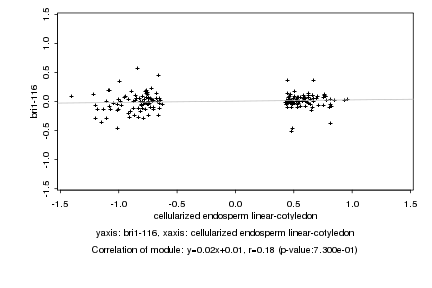
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cellularized endosperm linear-cotyledon |
Log2 signal ratio
bri1-116 |
| 1 |
267198_at |
AT2G30810
|
[Gibberellin-regulated family protein] |
0.953 |
0.038 |
| 2 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
0.930 |
0.031 |
| 3 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.842 |
0.024 |
| 4 |
253937_at |
AT4G26890
|
MAPKKK16, mitogen-activated protein kinase kinase kinase 16 |
0.821 |
-0.078 |
| 5 |
251857_at |
AT3G54770
|
ARP1, ABA-regulated RNA-binding protein 1 |
0.812 |
-0.362 |
| 6 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
0.808 |
0.049 |
| 7 |
264143_at |
AT1G79330
|
AMC6, ATMC5, metacaspase 5, ATMCP2b, metacaspase 2b, MC5, metacaspase 5, MCP2b, metacaspase 2b |
0.808 |
-0.046 |
| 8 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.800 |
-0.093 |
| 9 |
254003_at |
AT4G26290
|
unknown |
0.779 |
0.023 |
| 10 |
264361_at |
AT1G03300
|
unknown |
0.768 |
0.093 |
| 11 |
258571_at |
AT3G04420
|
anac048, NAC domain containing protein 48, NAC048, NAC domain containing protein 48 |
0.767 |
0.090 |
| 12 |
266387_at |
AT2G32360
|
[Ubiquitin-like superfamily protein] |
0.757 |
0.122 |
| 13 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.756 |
0.111 |
| 14 |
256435_at |
AT3G11180
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.750 |
-0.055 |
| 15 |
262396_at |
AT1G49470
|
unknown |
0.748 |
0.084 |
| 16 |
248553_at |
AT5G50170
|
[C2 calcium/lipid-binding and GRAM domain containing protein] |
0.710 |
-0.057 |
| 17 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.701 |
0.097 |
| 18 |
256292_at |
AT1G69430
|
unknown |
0.687 |
0.061 |
| 19 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.666 |
0.362 |
| 20 |
251822_at |
AT3G55060
|
unknown |
0.665 |
0.060 |
| 21 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
0.663 |
0.013 |
| 22 |
258442_at |
AT3G01015
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.660 |
0.111 |
| 23 |
254974_at |
AT4G10490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.659 |
0.112 |
| 24 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
0.652 |
-0.096 |
| 25 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.645 |
-0.152 |
| 26 |
251992_at |
AT3G53350
|
MIDD1, Microtubule Depletion Domain 1, RIP3, ROP interactive partner 3, RIP4, ROP interactive partner 4 |
0.639 |
0.042 |
| 27 |
263479_x_at |
AT2G04000
|
unknown |
0.637 |
-0.038 |
| 28 |
267521_at |
AT2G30480
|
unknown |
0.623 |
0.111 |
| 29 |
262036_at |
AT1G35530
|
At-FANCM, FANCM, Fanconi anemia complementation group M |
0.621 |
0.084 |
| 30 |
258130_at |
AT3G24510
|
[Defensin-like (DEFL) family protein] |
0.620 |
-0.031 |
| 31 |
261642_at |
AT1G27680
|
APL2, ADPGLC-PPase large subunit |
0.619 |
0.152 |
| 32 |
257542_at |
AT3G26050
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.609 |
-0.015 |
| 33 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
0.600 |
-0.083 |
| 34 |
245192_at |
AT1G67780
|
[Zinc-finger domain of monoamine-oxidase A repressor R1 protein] |
0.598 |
0.013 |
| 35 |
248471_at |
AT5G50840
|
unknown |
0.597 |
0.069 |
| 36 |
260471_at |
AT1G11070
|
unknown |
0.590 |
0.059 |
| 37 |
263130_at |
AT1G78650
|
POLD3 |
0.587 |
0.070 |
| 38 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
0.585 |
0.066 |
| 39 |
248359_at |
AT5G52410
|
unknown |
0.582 |
0.064 |
| 40 |
254720_at |
AT4G13610
|
MEE57, maternal effect embryo arrest 57 |
0.580 |
0.025 |
| 41 |
253347_at |
AT4G33610
|
[glycine-rich protein] |
0.576 |
0.120 |
| 42 |
252477_at |
AT3G46680
|
[UDP-Glycosyltransferase superfamily protein] |
0.568 |
0.017 |
| 43 |
259580_at |
AT1G28030
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.555 |
0.081 |
| 44 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.552 |
-0.075 |
| 45 |
248133_at |
AT5G54850
|
unknown |
0.544 |
0.012 |
| 46 |
256146_at |
AT1G48760
|
delta-ADR, delta-adaptin, PAT4, PROTEIN-AFFECTED TRAFFICKING 4 |
0.538 |
-0.020 |
| 47 |
248956_at |
AT5G45610
|
SUV2, SENSITIVE TO UV 2 |
0.538 |
0.080 |
| 48 |
264138_at |
AT1G78950
|
AtBAS, AtLUP4, BAS, beta-amyrin synthase |
0.531 |
0.073 |
| 49 |
259794_at |
AT1G64330
|
[myosin heavy chain-related] |
0.528 |
-0.089 |
| 50 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
0.527 |
-0.040 |
| 51 |
248279_at |
AT5G52910
|
ATIM, TIMELESS |
0.526 |
0.032 |
| 52 |
248379_at |
AT5G51700
|
ATRAR1, PBS2, PPHB SUSCEPTIBLE 2, RAR1, Required for Mla12 resistance 1, RPR2 |
0.523 |
0.016 |
| 53 |
261510_at |
AT1G71760
|
unknown |
0.516 |
0.044 |
| 54 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.502 |
0.182 |
| 55 |
253034_at |
AT4G38230
|
ATCPK26, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 26, CPK26, calcium-dependent protein kinase 26 |
0.500 |
-0.027 |
| 56 |
254463_at |
AT4G20280
|
TAF11, TBP-associated factor 11 |
0.498 |
0.090 |
| 57 |
253978_at |
AT4G26660
|
unknown |
0.489 |
0.066 |
| 58 |
252285_at |
AT3G49050
|
[alpha/beta-Hydrolases superfamily protein] |
0.488 |
-0.010 |
| 59 |
258504_at |
AT3G02590
|
[Fatty acid hydroxylase superfamily protein] |
0.487 |
-0.019 |
| 60 |
256040_at |
AT1G07270
|
[Cell division control, Cdc6] |
0.485 |
-0.044 |
| 61 |
250473_at |
AT5G10220
|
ANN6, annexin 6, ANNAT6, ANNEXIN ARABIDOPSIS THALIANA 6 |
0.484 |
0.009 |
| 62 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.483 |
-0.452 |
| 63 |
261840_at |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
0.478 |
0.029 |
| 64 |
266562_at |
AT2G23970
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.476 |
0.002 |
| 65 |
248252_at |
AT5G53250
|
AGP22, arabinogalactan protein 22, ATAGP22, ARABINOGALACTAN PROTEIN 22 |
0.475 |
-0.507 |
| 66 |
247187_at |
AT5G65490
|
unknown |
0.475 |
0.014 |
| 67 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
0.473 |
-0.097 |
| 68 |
248168_at |
AT5G54570
|
BGLU41, beta glucosidase 41 |
0.469 |
0.134 |
| 69 |
248955_at |
AT5G45600
|
GAS41, GLIOMAS 41, TAF14B, TBP-ASSOCIATED FACTOR 14B, YAF9a, homolog of yeast YAF9 a |
0.469 |
-0.016 |
| 70 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
0.465 |
-0.029 |
| 71 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
0.465 |
0.025 |
| 72 |
266639_at |
AT2G35520
|
DAD2, DEFENDER AGAINST CELL DEATH 2 |
0.456 |
0.041 |
| 73 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.455 |
0.090 |
| 74 |
253089_at |
AT4G36290
|
AtMORC1, CRT1, compromised recognition of TCV 1, MORC1, Microchidia 1 |
0.453 |
0.071 |
| 75 |
267135_at |
AT2G23430
|
ICK1, KRP1, KIP-RELATED PROTEIN 1 |
0.449 |
0.050 |
| 76 |
267218_at |
AT2G02515
|
unknown |
0.448 |
0.010 |
| 77 |
248432_at |
AT5G51390
|
unknown |
0.448 |
0.033 |
| 78 |
246261_at |
AT1G31810
|
AFH14, Formin Homology 14 |
0.445 |
-0.027 |
| 79 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.445 |
-0.089 |
| 80 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.442 |
0.016 |
| 81 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
0.438 |
0.153 |
| 82 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.438 |
0.367 |
| 83 |
246802_at |
AT5G27000
|
ATK4, kinesin 4, KATD, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA D |
0.437 |
-0.042 |
| 84 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
0.434 |
-0.039 |
| 85 |
257128_at |
AT3G20080
|
CYP705A15, cytochrome P450, family 705, subfamily A, polypeptide 15 |
0.430 |
0.024 |
| 86 |
264248_at |
AT1G78700
|
BEH4, BES1/BZR1 homolog 4 |
0.427 |
-0.001 |
| 87 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-1.411 |
0.099 |
| 88 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-1.223 |
0.122 |
| 89 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-1.207 |
-0.285 |
| 90 |
267122_at |
AT2G23550
|
ABE1, ALPHA/BETA FOLD HYDROLASE/ESTERASE 1, ATMES6, METHYL ESTERASE 6, MES6, methyl esterase 6 |
-1.201 |
-0.055 |
| 91 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-1.185 |
-0.121 |
| 92 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
-1.152 |
-0.350 |
| 93 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
-1.136 |
-0.123 |
| 94 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-1.113 |
-0.287 |
| 95 |
264557_at |
AT1G09550
|
[Pectinacetylesterase family protein] |
-1.107 |
0.008 |
| 96 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
-1.095 |
0.198 |
| 97 |
256599_at |
AT3G14760
|
unknown |
-1.084 |
0.206 |
| 98 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-1.080 |
-0.079 |
| 99 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-1.073 |
-0.133 |
| 100 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-1.047 |
-0.020 |
| 101 |
258091_at |
AT3G14560
|
unknown |
-1.014 |
-0.038 |
| 102 |
247827_at |
AT5G58500
|
LSH5, LIGHT SENSITIVE HYPOCOTYLS 5 |
-1.013 |
-0.151 |
| 103 |
261106_at |
AT1G62990
|
IXR11, KNAT7, KNOTTED-like homeobox of Arabidopsis thaliana 7 |
-1.012 |
-0.456 |
| 104 |
252138_at |
AT3G50990
|
PER36, peroxidase 36 |
-1.009 |
0.047 |
| 105 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
-1.006 |
-0.130 |
| 106 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.996 |
0.350 |
| 107 |
259124_at |
AT3G02310
|
AGL4, AGAMOUS-like 4, SEP2, SEPALLATA 2 |
-0.989 |
0.006 |
| 108 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
-0.983 |
-0.063 |
| 109 |
255014_at |
AT4G09960
|
AGL11, AGAMOUS-like 11, STK, SEEDSTICK |
-0.957 |
0.075 |
| 110 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.946 |
0.098 |
| 111 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.922 |
0.042 |
| 112 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.919 |
-0.203 |
| 113 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-0.913 |
-0.273 |
| 114 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.900 |
-0.168 |
| 115 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
-0.892 |
0.177 |
| 116 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.882 |
-0.106 |
| 117 |
249491_at |
AT5G39130
|
[RmlC-like cupins superfamily protein] |
-0.881 |
0.013 |
| 118 |
247214_at |
AT5G64850
|
unknown |
-0.872 |
-0.224 |
| 119 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
-0.863 |
0.105 |
| 120 |
248830_at |
AT5G47150
|
[YDG/SRA domain-containing protein] |
-0.860 |
0.020 |
| 121 |
256569_at |
AT3G19550
|
unknown |
-0.852 |
0.058 |
| 122 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.849 |
0.068 |
| 123 |
247800_at |
AT5G58570
|
unknown |
-0.844 |
0.577 |
| 124 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
-0.836 |
-0.110 |
| 125 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.835 |
-0.263 |
| 126 |
253056_at |
AT4G37650
|
SGR7, SHOOT GRAVITROPISM 7, SHR, SHORT ROOT |
-0.830 |
0.060 |
| 127 |
261813_at |
AT1G08280
|
[Glycosyltransferase family 29 (sialyltransferase) family protein] |
-0.817 |
-0.159 |
| 128 |
246990_at |
AT5G67360
|
ARA12 |
-0.815 |
0.012 |
| 129 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
-0.813 |
-0.043 |
| 130 |
260490_at |
AT1G51500
|
ABCG12, ATP-binding cassette G12, AtABCG12, ATWBC12, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX 12, CER5, ECERIFERUM 5, D3, WBC12, WHITE-BROWN COMPLEX 12 |
-0.807 |
-0.065 |
| 131 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.803 |
0.009 |
| 132 |
266444_at |
AT2G43150
|
[Proline-rich extensin-like family protein] |
-0.802 |
0.098 |
| 133 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
-0.799 |
-0.108 |
| 134 |
254077_at |
AT4G25640
|
ATDTX35, detoxifying efflux carrier 35, DTX35, detoxifying efflux carrier 35, FFT, FLOWER FLAVONOID TRANSPORTER |
-0.796 |
-0.042 |
| 135 |
266640_at |
AT2G35585
|
unknown |
-0.790 |
-0.283 |
| 136 |
253053_at |
AT4G37470
|
KAI2, KARRIKIN INSENSITIVE 2 |
-0.780 |
0.042 |
| 137 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.780 |
-0.016 |
| 138 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.777 |
0.186 |
| 139 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.776 |
-0.126 |
| 140 |
267525_at |
AT2G30560
|
[glycine-rich protein] |
-0.770 |
0.075 |
| 141 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.769 |
0.193 |
| 142 |
246041_at |
AT5G19290
|
[alpha/beta-Hydrolases superfamily protein] |
-0.768 |
-0.026 |
| 143 |
249479_at |
AT5G38960
|
[RmlC-like cupins superfamily protein] |
-0.766 |
0.144 |
| 144 |
253052_at |
AT4G37310
|
CYP81H1, cytochrome P450, family 81, subfamily H, polypeptide 1 |
-0.762 |
0.164 |
| 145 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.761 |
-0.044 |
| 146 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.761 |
0.127 |
| 147 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.751 |
0.080 |
| 148 |
261921_at |
AT1G65900
|
unknown |
-0.750 |
-0.036 |
| 149 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
-0.748 |
0.011 |
| 150 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.747 |
-0.224 |
| 151 |
262539_at |
AT1G17200
|
[Uncharacterised protein family (UPF0497)] |
-0.746 |
0.067 |
| 152 |
256865_at |
AT3G23820
|
GAE6, UDP-D-glucuronate 4-epimerase 6 |
-0.743 |
-0.023 |
| 153 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.735 |
0.060 |
| 154 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.731 |
-0.097 |
| 155 |
267124_at |
AT2G23570
|
ATMES19, ARABIDOPSIS THALIANA METHYL ESTERASE 19, MES19, methyl esterase 19 |
-0.727 |
0.046 |
| 156 |
261181_at |
AT1G34580
|
[Major facilitator superfamily protein] |
-0.720 |
0.227 |
| 157 |
250690_at |
AT5G06530
|
ABCG22, ATP-binding cassette G22, AtABCG22, Arabidopsis thaliana ATP-binding cassette G22 |
-0.719 |
0.002 |
| 158 |
249750_at |
AT5G24570
|
unknown |
-0.717 |
0.030 |
| 159 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.709 |
-0.086 |
| 160 |
252050_at |
AT3G52550
|
unknown |
-0.708 |
0.034 |
| 161 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.704 |
-0.127 |
| 162 |
245375_at |
AT4G17660
|
[Protein kinase superfamily protein] |
-0.685 |
0.052 |
| 163 |
267093_at |
AT2G38170
|
ATCAX1, CAX1, cation exchanger 1, RCI4, RARE COLD INDUCIBLE 4 |
-0.679 |
0.148 |
| 164 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.674 |
0.017 |
| 165 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.667 |
-0.224 |
| 166 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.660 |
0.454 |
| 167 |
250920_at |
AT5G03390
|
unknown |
-0.659 |
-0.029 |
| 168 |
260917_at |
AT1G02700
|
unknown |
-0.659 |
0.058 |
| 169 |
254594_at |
AT4G18930
|
[RNA ligase/cyclic nucleotide phosphodiesterase family protein] |
-0.658 |
-0.108 |
| 170 |
251189_at |
AT3G62650
|
unknown |
-0.645 |
0.029 |
| 171 |
255056_at |
AT4G09820
|
AtTT8, BHLH42, TT8, TRANSPARENT TESTA 8 |
-0.633 |
-0.036 |