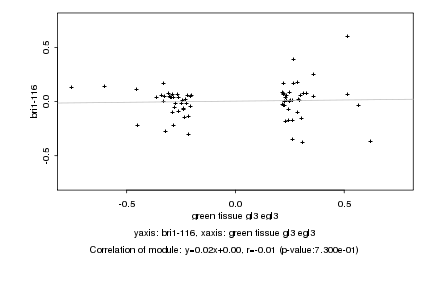
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
green tissue gl3 egl3 |
Log2 signal ratio
bri1-116 |
| 1 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.620 |
-0.361 |
| 2 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.565 |
-0.034 |
| 3 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
0.514 |
0.607 |
| 4 |
266837_x_at |
AT2G25990
|
unknown |
0.513 |
0.068 |
| 5 |
250201_at |
AT5G14230
|
unknown |
0.355 |
0.253 |
| 6 |
249536_at |
AT5G38760
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.355 |
0.054 |
| 7 |
247732_at |
AT5G59600
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.323 |
0.077 |
| 8 |
260265_at |
AT1G68510
|
LBD42, LOB domain-containing protein 42 |
0.311 |
0.075 |
| 9 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
0.307 |
-0.371 |
| 10 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.303 |
-0.153 |
| 11 |
260917_at |
AT1G02700
|
unknown |
0.298 |
0.058 |
| 12 |
255945_at |
AT5G28610
|
unknown |
0.291 |
0.016 |
| 13 |
254813_at |
AT4G12350
|
AtMYB42, myb domain protein 42, MYB42, myb domain protein 42 |
0.286 |
0.021 |
| 14 |
245972_at |
AT5G20680
|
TBL16, TRICHOME BIREFRINGENCE-LIKE 16 |
0.283 |
-0.101 |
| 15 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.283 |
0.182 |
| 16 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.264 |
0.397 |
| 17 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
0.263 |
0.171 |
| 18 |
257331_at |
ATMG01330
|
ORF107H |
0.262 |
0.018 |
| 19 |
253165_at |
AT4G35320
|
unknown |
0.261 |
-0.347 |
| 20 |
254294_at |
AT4G23070
|
ATRBL7, RHOMBOID-like protein 7, RBL7, RHOMBOID-like protein 7 |
0.259 |
-0.168 |
| 21 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
0.250 |
0.010 |
| 22 |
250164_at |
AT5G15280
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.248 |
0.089 |
| 23 |
248490_at |
AT5G50940
|
[RNA-binding KH domain-containing protein] |
0.247 |
0.007 |
| 24 |
249072_at |
AT5G44060
|
unknown |
0.242 |
-0.175 |
| 25 |
258576_at |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.241 |
-0.071 |
| 26 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.234 |
0.064 |
| 27 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.232 |
0.014 |
| 28 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.230 |
0.041 |
| 29 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
0.226 |
-0.182 |
| 30 |
247068_at |
AT5G66800
|
unknown |
0.222 |
-0.032 |
| 31 |
249693_at |
AT5G35750
|
AHK2, histidine kinase 2, HK2, histidine kinase 2 |
0.221 |
0.004 |
| 32 |
251711_at |
AT3G56960
|
PIP5K4, phosphatidyl inositol monophosphate 5 kinase 4 |
0.220 |
0.173 |
| 33 |
249223_at |
AT5G42120
|
LecRK-S.6, L-type lectin receptor kinase S.6 |
0.219 |
-0.030 |
| 34 |
266470_at |
AT2G31080
|
[non-LTR retrotransposon family (LINE), has a 1.3e-49 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
0.218 |
0.071 |
| 35 |
253669_at |
AT4G30000
|
[Dihydropterin pyrophosphokinase / Dihydropteroate synthase] |
0.218 |
0.078 |
| 36 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
0.216 |
-0.025 |
| 37 |
255191_at |
AT4G07370
|
[gypsy-like retrotransposon family (Athila), has a 1.7e-75 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.215 |
0.090 |
| 38 |
267459_at |
AT2G33850
|
unknown |
-0.757 |
0.138 |
| 39 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-0.602 |
0.145 |
| 40 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.457 |
0.116 |
| 41 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.451 |
-0.215 |
| 42 |
258802_at |
AT3G04650
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.367 |
0.044 |
| 43 |
248479_at |
AT5G50910
|
unknown |
-0.342 |
0.064 |
| 44 |
255900_at |
AT1G17830
|
unknown |
-0.333 |
0.172 |
| 45 |
257992_at |
AT3G19880
|
[F-box and associated interaction domains-containing protein] |
-0.331 |
0.001 |
| 46 |
266023_at |
AT2G05930
|
[copia-like retrotransposon family, has a 0. P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.327 |
0.048 |
| 47 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.323 |
-0.268 |
| 48 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.311 |
0.081 |
| 49 |
248829_at |
AT5G47130
|
[Bax inhibitor-1 family protein] |
-0.304 |
0.050 |
| 50 |
255493_at |
AT4G02690
|
AtLFG3, LFG3, LIFEGUARD 3 |
-0.301 |
0.042 |
| 51 |
254069_at |
AT4G25434
|
ATNUDT10, nudix hydrolase homolog 10, NUDT10, nudix hydrolase homolog 10 |
-0.296 |
0.044 |
| 52 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.291 |
-0.100 |
| 53 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.291 |
0.066 |
| 54 |
261736_at |
AT1G47810
|
[F-box and associated interaction domains-containing protein] |
-0.289 |
0.044 |
| 55 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
-0.285 |
-0.221 |
| 56 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-0.282 |
-0.032 |
| 57 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.276 |
-0.012 |
| 58 |
260256_at |
AT1G74350
|
[Intron maturase, type II family protein] |
-0.270 |
0.068 |
| 59 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
-0.264 |
-0.083 |
| 60 |
255347_at |
AT4G03810
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:BAA22288 pol polyprotein (Ty1_Copia-element) (Oryza australiensis)GB:BAA22288 polyprotein (Ty1_Copia-element) (Oryza australiensis)gi|2443320|dbj|BAA22288.1| polyprotein (RIRE1) (Oryza australiensis) (Ty1_Copia-element)] |
-0.263 |
0.041 |
| 61 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.252 |
-0.018 |
| 62 |
246437_at |
AT5G17540
|
[HXXXD-type acyl-transferase family protein] |
-0.245 |
0.014 |
| 63 |
255125_at |
AT4G08250
|
[GRAS family transcription factor] |
-0.243 |
-0.057 |
| 64 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.239 |
-0.069 |
| 65 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.235 |
-0.139 |
| 66 |
257416_at |
AT2G17750
|
NIP1, NEP-interacting protein 1 |
-0.233 |
0.019 |
| 67 |
260945_at |
AT1G05950
|
unknown |
-0.229 |
-0.010 |
| 68 |
263850_at |
AT2G04480
|
unknown |
-0.223 |
0.063 |
| 69 |
263013_at |
AT1G23380
|
KNAT6, KNOTTED1-like homeobox gene 6, KNAT6L, KNAT6S |
-0.220 |
-0.301 |
| 70 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.219 |
-0.131 |
| 71 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
-0.210 |
-0.039 |
| 72 |
267205_at |
AT2G30820
|
unknown |
-0.207 |
0.054 |
| 73 |
256546_at |
AT3G14820
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.206 |
0.060 |