|
probeID |
AGICode |
Annotation |
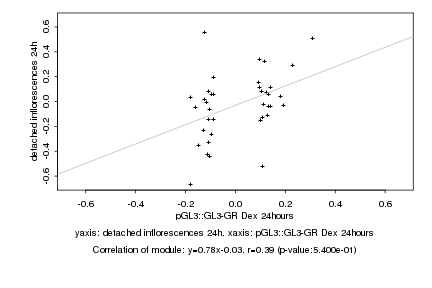
Log2 signal ratio
pGL3::GL3-GR Dex 24hours |
Log2 signal ratio
detached inflorescences 24h |
| 1 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.307 |
0.513 |
| 2 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.228 |
0.294 |
| 3 |
248355_at |
AT5G52340
|
ATEXO70A2, exocyst subunit exo70 family protein A2, EXO70A2, exocyst subunit exo70 family protein A2 |
0.190 |
-0.027 |
| 4 |
260963_at |
AT1G44990
|
unknown |
0.177 |
0.043 |
| 5 |
246686_at |
AT5G33380
|
unknown |
0.138 |
0.117 |
| 6 |
245153_at |
AT5G12450
|
[FBD-like domain family protein] |
0.137 |
-0.034 |
| 7 |
253027_at |
AT4G38150
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.131 |
-0.035 |
| 8 |
251385_at |
AT3G60790
|
[F-box family protein] |
0.130 |
0.062 |
| 9 |
263427_at |
AT2G22260
|
ALKBH2, homolog of E. coli alkB |
0.126 |
-0.110 |
| 10 |
261533_at |
AT1G71690
|
unknown |
0.123 |
0.073 |
| 11 |
247905_at |
AT5G57400
|
unknown |
0.114 |
0.328 |
| 12 |
258343_at |
AT3G22810
|
[FUNCTIONS IN: phosphoinositide binding] |
0.110 |
-0.021 |
| 13 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
0.106 |
-0.521 |
| 14 |
261425_at |
AT1G18880
|
AtNPF2.9, NPF2.9, NRT1/ PTR family 2.9, NRT1.9, nitrate transporter 1.9 |
0.105 |
-0.122 |
| 15 |
255339_at |
AT4G04480
|
unknown |
0.103 |
0.086 |
| 16 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
0.098 |
-0.145 |
| 17 |
263757_at |
AT2G21430
|
[Papain family cysteine protease] |
0.094 |
0.116 |
| 18 |
247061_at |
AT5G66780
|
unknown |
0.093 |
0.342 |
| 19 |
250624_at |
AT5G07330
|
unknown |
0.090 |
0.161 |
| 20 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.183 |
-0.660 |
| 21 |
250956_at |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
-0.182 |
0.039 |
| 22 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.164 |
-0.044 |
| 23 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.151 |
-0.347 |
| 24 |
266721_at |
AT2G03220
|
ATFT1, ARABIDOPSIS THALIANA FUCOSYLTRANSFERASE 1, ATFUT1, ARABIDOPSIS THALIANA FUCOSYLTRANSFERASE 1, FT1, fucosyltransferase 1, MUR2, MURUS 2 |
-0.130 |
-0.230 |
| 25 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.128 |
0.556 |
| 26 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.127 |
0.022 |
| 27 |
259566_at |
AT1G20520
|
unknown |
-0.118 |
-0.001 |
| 28 |
249167_at |
AT5G42860
|
unknown |
-0.116 |
-0.423 |
| 29 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.111 |
0.082 |
| 30 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.110 |
-0.141 |
| 31 |
264600_at |
AT1G04730
|
CTF18, CHROMOSOME TRANSMISSION FIDELITY 18 |
-0.109 |
-0.323 |
| 32 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.107 |
-0.439 |
| 33 |
250789_at |
AT5G05630
|
PUT3, POLYAMINE UPTAKE TRANSPORTER 3, RMV1, resistant to methyl viologen 1 |
-0.106 |
-0.064 |
| 34 |
267192_at |
AT2G30890
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
-0.097 |
-0.261 |
| 35 |
267137_at |
AT2G23410
|
ACPT, cis-prenyltransferase, AtcPT3, CPT, cis-prenyltransferase, cPT3, cis-prenyltransferase 3 |
-0.097 |
0.059 |
| 36 |
250026_at |
AT5G18090
|
[AP2/B3-like transcriptional factor family protein] |
-0.091 |
-0.138 |
| 37 |
266749_at |
AT2G47060
|
PTI1-4, Pto-interacting 1-4 |
-0.088 |
0.063 |
| 38 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
-0.088 |
0.198 |