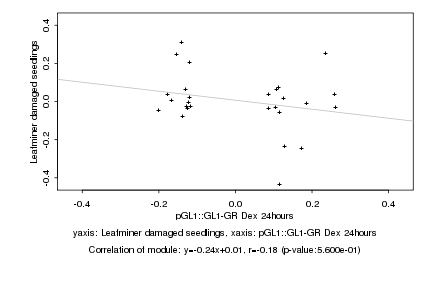
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
pGL1::GL1-GR Dex 24hours |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
0.261 |
-0.028 |
| 2 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.258 |
0.039 |
| 3 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.234 |
0.254 |
| 4 |
262033_at |
AT1G37140
|
MCT1, MEI2 C-terminal RRM only like 1 |
0.184 |
-0.007 |
| 5 |
261208_at |
AT1G12930
|
[ARM repeat superfamily protein] |
0.171 |
-0.243 |
| 6 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
0.128 |
-0.233 |
| 7 |
261720_at |
AT1G08460
|
ATHDA8, HDA08, histone deacetylase 8, HDA8, HISTONE DEACETYLASE 8 |
0.124 |
0.021 |
| 8 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.115 |
-0.055 |
| 9 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
0.113 |
-0.430 |
| 10 |
253400_at |
AT4G32860
|
unknown |
0.112 |
0.077 |
| 11 |
249069_at |
AT5G44010
|
unknown |
0.105 |
0.068 |
| 12 |
264528_at |
AT1G30810
|
JMJ18, Jumonji domain-containing protein 18 |
0.102 |
-0.031 |
| 13 |
259285_at |
AT3G11460
|
MEF10, mitochondrial RNA editing factor 10 |
0.086 |
-0.035 |
| 14 |
247885_at |
AT5G57830
|
unknown |
0.085 |
0.038 |
| 15 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
-0.201 |
-0.042 |
| 16 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.179 |
0.042 |
| 17 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
-0.168 |
0.009 |
| 18 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.156 |
0.248 |
| 19 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-0.141 |
0.311 |
| 20 |
256507_at |
AT1G75150
|
unknown |
-0.139 |
-0.078 |
| 21 |
248675_at |
AT5G48820
|
ICK6, inhibitor/interactor with cyclin-dependent kinase, KRP3, KIP-RELATED PROTEIN 3 |
-0.133 |
0.064 |
| 22 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.129 |
-0.022 |
| 23 |
254155_at |
AT4G24480
|
[Protein kinase superfamily protein] |
-0.127 |
-0.036 |
| 24 |
262915_at |
AT1G59940
|
ARR3, response regulator 3 |
-0.124 |
-0.003 |
| 25 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
-0.122 |
0.207 |
| 26 |
266762_at |
AT2G47120
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.120 |
0.022 |
| 27 |
253211_at |
AT4G34880
|
[Amidase family protein] |
-0.119 |
-0.026 |