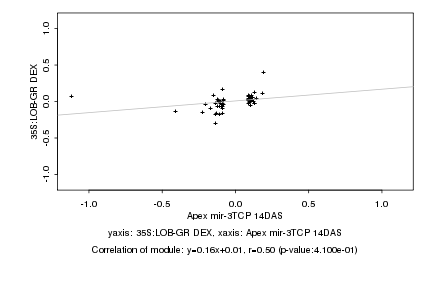
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Apex mir-3TCP 14DAS |
Log2 signal ratio
35S:LOB-GR DEX |
| 1 |
261684_at |
AT1G47400
|
unknown |
0.191 |
0.399 |
| 2 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.182 |
0.118 |
| 3 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
0.142 |
0.049 |
| 4 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.127 |
0.129 |
| 5 |
264987_at |
AT1G27030
|
unknown |
0.125 |
-0.021 |
| 6 |
263535_at |
AT2G24970
|
unknown |
0.125 |
-0.019 |
| 7 |
254800_at |
AT4G13070
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
0.121 |
0.007 |
| 8 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
0.115 |
0.061 |
| 9 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.113 |
0.041 |
| 10 |
266406_at |
AT2G38570
|
unknown |
0.112 |
0.067 |
| 11 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.107 |
0.058 |
| 12 |
266567_at |
AT2G24050
|
eIFiso4G2, eukaryotic translation Initiation Factor isoform 4G2 |
0.105 |
0.085 |
| 13 |
250388_at |
AT5G11310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.102 |
0.022 |
| 14 |
260090_at |
AT1G73310
|
scpl4, serine carboxypeptidase-like 4 |
0.102 |
0.009 |
| 15 |
262330_at |
AT1G64070
|
RLM1, RESISTANCE TO LEPTOSPHAERIA MACULANS 1 |
0.096 |
-0.051 |
| 16 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
0.095 |
-0.001 |
| 17 |
252360_at |
AT3G48480
|
[Cysteine proteinases superfamily protein] |
0.095 |
0.052 |
| 18 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
0.095 |
0.002 |
| 19 |
257498_at |
AT1G69660
|
[TRAF-like family protein] |
0.093 |
0.044 |
| 20 |
253479_at |
AT4G32360
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
0.093 |
0.080 |
| 21 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
0.093 |
0.078 |
| 22 |
263415_at |
AT2G17250
|
EMB2762, EMBRYO DEFECTIVE 2762, NOC4, NucleOlar Complex associated 4 |
0.090 |
0.031 |
| 23 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.089 |
0.023 |
| 24 |
266903_at |
AT2G34570
|
MEE21, maternal effect embryo arrest 21 |
0.089 |
0.038 |
| 25 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
0.089 |
-0.022 |
| 26 |
254581_at |
AT4G19440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.089 |
0.020 |
| 27 |
267148_at |
AT2G23470
|
RUS4, ROOT UV-B SENSITIVE 4 |
0.088 |
0.054 |
| 28 |
247606_at |
AT5G61000
|
ATRPA70D, RPA70D |
0.087 |
0.029 |
| 29 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
0.086 |
0.096 |
| 30 |
254901_at |
AT4G11530
|
CRK34, cysteine-rich RLK (RECEPTOR-like protein kinase) 34 |
0.085 |
0.082 |
| 31 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-1.124 |
0.082 |
| 32 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.411 |
-0.133 |
| 33 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.229 |
-0.146 |
| 34 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.210 |
-0.031 |
| 35 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.176 |
-0.087 |
| 36 |
247072_at |
AT5G66490
|
unknown |
-0.154 |
0.086 |
| 37 |
263233_at |
AT1G05577
|
unknown |
-0.140 |
-0.018 |
| 38 |
247585_at |
AT5G60680
|
unknown |
-0.140 |
-0.176 |
| 39 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
-0.138 |
-0.295 |
| 40 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
-0.132 |
-0.151 |
| 41 |
260403_at |
AT1G69810
|
ATWRKY36, WRKY36, WRKY DNA-binding protein 36 |
-0.127 |
0.040 |
| 42 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.126 |
-0.060 |
| 43 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.122 |
0.024 |
| 44 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.117 |
0.003 |
| 45 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.115 |
-0.171 |
| 46 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.111 |
-0.061 |
| 47 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
-0.109 |
0.014 |
| 48 |
258282_at |
AT3G26910
|
[hydroxyproline-rich glycoprotein family protein] |
-0.108 |
-0.014 |
| 49 |
262789_at |
AT1G10810
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.102 |
0.008 |
| 50 |
255221_at |
AT4G05150
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.097 |
-0.065 |
| 51 |
252906_at |
AT4G39640
|
GGT1, gamma-glutamyl transpeptidase 1 |
-0.095 |
-0.152 |
| 52 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
-0.093 |
-0.060 |
| 53 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.093 |
-0.095 |
| 54 |
250877_at |
AT5G04040
|
SDP1, SUGAR-DEPENDENT1 |
-0.092 |
-0.039 |
| 55 |
246975_at |
AT5G24890
|
unknown |
-0.091 |
-0.022 |
| 56 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.090 |
0.173 |
| 57 |
253588_at |
AT4G30790
|
[INVOLVED IN: autophagy] |
-0.087 |
-0.037 |
| 58 |
263858_at |
AT2G04370
|
unknown |
-0.087 |
0.032 |
| 59 |
267424_at |
AT2G34800
|
unknown |
-0.086 |
0.019 |