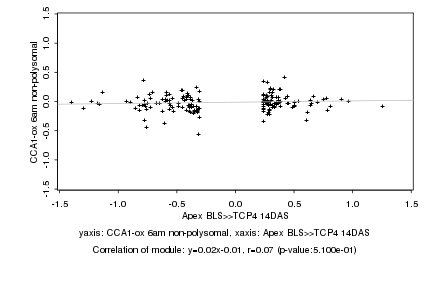
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Apex BLS>>TCP4 14DAS |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
255471_at |
AT4G03050
|
AOP3 |
1.248 |
-0.073 |
| 2 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
0.962 |
0.017 |
| 3 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
0.902 |
0.039 |
| 4 |
250135_at |
AT5G15360
|
unknown |
0.809 |
-0.083 |
| 5 |
254566_at |
AT4G19240
|
unknown |
0.781 |
-0.149 |
| 6 |
262421_at |
AT1G50290
|
unknown |
0.773 |
0.066 |
| 7 |
253689_at |
AT4G29770
|
unknown |
0.750 |
0.046 |
| 8 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.697 |
-0.012 |
| 9 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.663 |
0.087 |
| 10 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.646 |
-0.023 |
| 11 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.640 |
0.027 |
| 12 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
0.638 |
-0.068 |
| 13 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.612 |
-0.174 |
| 14 |
249656_at |
AT5G37130
|
[Protein prenylyltransferase superfamily protein] |
0.605 |
-0.317 |
| 15 |
266808_at |
AT2G29995
|
unknown |
0.538 |
0.014 |
| 16 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.501 |
-0.003 |
| 17 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
0.499 |
-0.060 |
| 18 |
262160_at |
AT1G52590
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.498 |
-0.075 |
| 19 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
0.485 |
-0.093 |
| 20 |
256599_at |
AT3G14760
|
unknown |
0.447 |
-0.031 |
| 21 |
260148_at |
AT1G52800
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.438 |
0.096 |
| 22 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
0.436 |
-0.017 |
| 23 |
253532_at |
AT4G31570
|
unknown |
0.423 |
0.055 |
| 24 |
249510_at |
AT5G38510
|
[Rhomboid-related intramembrane serine protease family protein] |
0.411 |
0.418 |
| 25 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.384 |
-0.080 |
| 26 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
0.379 |
0.208 |
| 27 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
0.376 |
0.002 |
| 28 |
255028_at |
AT4G09890
|
unknown |
0.368 |
0.222 |
| 29 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.368 |
-0.011 |
| 30 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.365 |
-0.025 |
| 31 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.364 |
0.027 |
| 32 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.362 |
0.078 |
| 33 |
263688_at |
AT1G26920
|
unknown |
0.345 |
-0.005 |
| 34 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
0.345 |
0.069 |
| 35 |
260367_at |
AT1G69760
|
unknown |
0.344 |
-0.046 |
| 36 |
255940_at |
AT1G20380
|
[Prolyl oligopeptidase family protein] |
0.342 |
0.078 |
| 37 |
248148_at |
AT5G54930
|
[AT hook motif-containing protein] |
0.333 |
-0.095 |
| 38 |
254287_at |
AT4G22960
|
unknown |
0.331 |
-0.033 |
| 39 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.331 |
-0.077 |
| 40 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
0.317 |
0.208 |
| 41 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
0.313 |
0.072 |
| 42 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.310 |
-0.053 |
| 43 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
0.309 |
-0.086 |
| 44 |
254946_at |
AT4G10950
|
[SGNH hydrolase-type esterase superfamily protein] |
0.309 |
0.168 |
| 45 |
244997_at |
ATCG00170
|
RPOC2 |
0.306 |
0.116 |
| 46 |
247617_at |
AT5G60270
|
LecRK-I.7, L-type lectin receptor kinase I.7 |
0.306 |
0.020 |
| 47 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.298 |
0.224 |
| 48 |
266402_at |
AT2G38780
|
unknown |
0.297 |
0.017 |
| 49 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.294 |
-0.042 |
| 50 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
0.294 |
0.219 |
| 51 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
0.290 |
-0.142 |
| 52 |
260843_at |
AT1G29060
|
[Target SNARE coiled-coil domain protein] |
0.288 |
-0.220 |
| 53 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
0.286 |
0.166 |
| 54 |
249148_at |
AT5G43260
|
[chaperone protein dnaJ-related] |
0.286 |
-0.121 |
| 55 |
254901_at |
AT4G11530
|
CRK34, cysteine-rich RLK (RECEPTOR-like protein kinase) 34 |
0.281 |
-0.188 |
| 56 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.279 |
-0.053 |
| 57 |
263115_at |
AT1G03055
|
AtD27, A. thaliana homolog of rice D27, D27, DWARF27 |
0.277 |
-0.003 |
| 58 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
0.273 |
0.330 |
| 59 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.272 |
0.101 |
| 60 |
262921_at |
AT1G79430
|
APL, ALTERED PHLOEM DEVELOPMENT, WDY, WOODY |
0.272 |
-0.214 |
| 61 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.271 |
0.018 |
| 62 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.270 |
-0.050 |
| 63 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.270 |
-0.048 |
| 64 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
0.270 |
0.007 |
| 65 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.265 |
-0.027 |
| 66 |
264909_at |
AT2G17300
|
unknown |
0.263 |
0.109 |
| 67 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
0.263 |
0.072 |
| 68 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.256 |
-0.013 |
| 69 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
0.256 |
-0.013 |
| 70 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.255 |
0.009 |
| 71 |
263177_at |
AT1G05540
|
unknown |
0.255 |
-0.007 |
| 72 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.250 |
0.023 |
| 73 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
0.248 |
0.025 |
| 74 |
257881_at |
AT3G17180
|
scpl33, serine carboxypeptidase-like 33 |
0.242 |
0.032 |
| 75 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
0.239 |
-0.340 |
| 76 |
245020_at |
ATCG00540
|
PETA, photosynthetic electron transfer A |
0.238 |
0.112 |
| 77 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.238 |
0.348 |
| 78 |
259134_at |
AT3G05390
|
unknown |
0.237 |
-0.118 |
| 79 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
0.237 |
-0.074 |
| 80 |
260232_at |
AT1G74640
|
[alpha/beta-Hydrolases superfamily protein] |
0.236 |
0.039 |
| 81 |
247763_at |
AT5G59180
|
NRPB7 |
0.236 |
0.129 |
| 82 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.236 |
0.002 |
| 83 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
0.235 |
-0.039 |
| 84 |
259683_at |
AT1G63050
|
AtLPLAT2, LPCAT2, lysophosphatidylcholine acyltransferase 2, LPLAT2, lysophospholipid acyltransferase 2 |
0.232 |
-0.008 |
| 85 |
266567_at |
AT2G24050
|
eIFiso4G2, eukaryotic translation Initiation Factor isoform 4G2 |
0.232 |
-0.155 |
| 86 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
0.232 |
-0.128 |
| 87 |
248348_at |
AT5G52190
|
[Sugar isomerase (SIS) family protein] |
0.232 |
-0.005 |
| 88 |
267459_at |
AT2G33850
|
unknown |
-1.405 |
-0.016 |
| 89 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.298 |
0.010 |
| 90 |
256021_at |
AT1G58270
|
ZW9 |
-1.236 |
0.006 |
| 91 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-1.184 |
-0.031 |
| 92 |
262010_at |
AT1G35612
|
[expressed protein] |
-1.165 |
-0.034 |
| 93 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-1.137 |
0.162 |
| 94 |
265611_at |
AT2G25510
|
unknown |
-0.938 |
0.007 |
| 95 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.898 |
-0.004 |
| 96 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.856 |
-0.114 |
| 97 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.844 |
0.079 |
| 98 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-0.823 |
-0.139 |
| 99 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.821 |
-0.062 |
| 100 |
257221_at |
AT3G27920
|
ATGL1, ATMYB0, myb domain protein 0, GL1, GLABRA 1, MYB0, myb domain protein 0 |
-0.797 |
-0.064 |
| 101 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.790 |
0.369 |
| 102 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.782 |
-0.040 |
| 103 |
248079_at |
AT5G55790
|
unknown |
-0.781 |
0.028 |
| 104 |
258424_at |
AT3G16750
|
unknown |
-0.777 |
-0.315 |
| 105 |
252659_at |
AT3G44430
|
unknown |
-0.769 |
-0.070 |
| 106 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.766 |
-0.432 |
| 107 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.760 |
-0.132 |
| 108 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
-0.756 |
-0.025 |
| 109 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.735 |
0.133 |
| 110 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.730 |
-0.091 |
| 111 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
-0.730 |
0.056 |
| 112 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.710 |
0.155 |
| 113 |
248016_at |
AT5G56380
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.679 |
-0.024 |
| 114 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.677 |
-0.026 |
| 115 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.656 |
-0.020 |
| 116 |
260883_at |
AT1G29270
|
unknown |
-0.626 |
0.046 |
| 117 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.624 |
-0.163 |
| 118 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
-0.608 |
-0.365 |
| 119 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.605 |
0.003 |
| 120 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
-0.604 |
0.037 |
| 121 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
-0.591 |
0.119 |
| 122 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.590 |
0.157 |
| 123 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.583 |
0.032 |
| 124 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
-0.568 |
-0.122 |
| 125 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.568 |
0.125 |
| 126 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
-0.564 |
0.020 |
| 127 |
246079_at |
AT5G20450
|
unknown |
-0.563 |
-0.031 |
| 128 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.559 |
-0.063 |
| 129 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.540 |
-0.090 |
| 130 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.539 |
0.066 |
| 131 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.536 |
-0.163 |
| 132 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
-0.492 |
-0.076 |
| 133 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.489 |
-0.026 |
| 134 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.462 |
0.195 |
| 135 |
259656_at |
AT1G55200
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.460 |
0.203 |
| 136 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.458 |
0.068 |
| 137 |
267077_at |
AT2G40970
|
MYBC1 |
-0.458 |
-0.086 |
| 138 |
255121_at |
AT4G08480
|
MAPKKK9, mitogen-activated protein kinase kinase kinase 9, MEKK2, MAPK/ERK KINASE KINASE 2, SUMM1, suppressor of mkk1 mkk2 1 |
-0.445 |
0.093 |
| 139 |
258000_at |
AT3G28940
|
[AIG2-like (avirulence induced gene) family protein] |
-0.435 |
0.021 |
| 140 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
-0.425 |
-0.145 |
| 141 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
-0.424 |
0.078 |
| 142 |
259281_at |
AT3G01140
|
AtMYB106, myb domain protein 106, MYB106, myb domain protein 106, NOK, NOECK |
-0.422 |
0.041 |
| 143 |
247934_at |
AT5G57000
|
unknown |
-0.412 |
0.150 |
| 144 |
245952_at |
AT5G28500
|
unknown |
-0.411 |
0.110 |
| 145 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
-0.404 |
0.112 |
| 146 |
245055_at |
AT2G26470
|
unknown |
-0.404 |
-0.059 |
| 147 |
246393_at |
AT1G58150
|
unknown |
-0.399 |
-0.093 |
| 148 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.398 |
-0.163 |
| 149 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
-0.397 |
-0.071 |
| 150 |
260515_at |
AT1G51460
|
ABCG13, ATP-binding cassette G13 |
-0.395 |
-0.055 |
| 151 |
251165_at |
AT3G63340
|
[Protein phosphatase 2C family protein] |
-0.390 |
0.083 |
| 152 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.390 |
-0.180 |
| 153 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.386 |
0.048 |
| 154 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.383 |
-0.100 |
| 155 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.375 |
-0.034 |
| 156 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.368 |
0.034 |
| 157 |
248054_at |
AT5G55820
|
WYR, WYRD |
-0.362 |
-0.083 |
| 158 |
260153_at |
AT1G52760
|
LysoPL2, lysophospholipase 2 |
-0.359 |
-0.189 |
| 159 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.356 |
-0.153 |
| 160 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
-0.352 |
-0.154 |
| 161 |
249331_at |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
-0.338 |
0.252 |
| 162 |
247124_at |
AT5G66060
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.338 |
-0.084 |
| 163 |
267054_at |
AT2G38370
|
unknown |
-0.337 |
-0.139 |
| 164 |
252654_at |
AT3G44740
|
[Class II aaRS and biotin synthetases superfamily protein] |
-0.329 |
-0.138 |
| 165 |
247885_at |
AT5G57830
|
unknown |
-0.325 |
-0.173 |
| 166 |
256493_at |
AT1G31600
|
AtTRM9, Arabidopsis thaliana tRNA methyltransferase 9, TRM9, tRNA methyltransferase 9 |
-0.325 |
-0.164 |
| 167 |
264459_at |
AT1G10160
|
[non-LTR retrotransposon family (LINE), has a 3.9e-39 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
-0.321 |
-0.108 |
| 168 |
262477_at |
AT1G11220
|
unknown |
-0.321 |
0.047 |
| 169 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.316 |
-0.561 |
| 170 |
248539_at |
AT5G50130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.312 |
0.182 |
| 171 |
253363_at |
AT4G33130
|
unknown |
-0.312 |
0.017 |
| 172 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.311 |
-0.112 |
| 173 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
-0.308 |
-0.270 |