|
probeID |
AGICode |
Annotation |
Log2 signal ratio
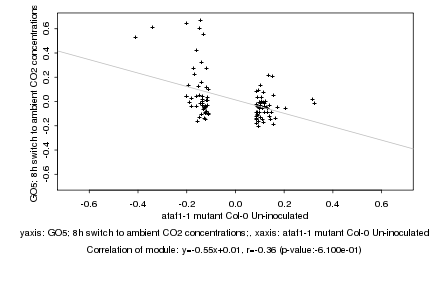
ataf1-1 mutant Col-0 Un-inoculated |
Log2 signal ratio
GO5; 8h switch to ambient CO2 concentrations; |
| 1 |
248304_at |
AT5G53180
|
ATPTB2, POLYPYRIMIDINE TRACT-BINDING PROTEIN 2, PTB2, polypyrimidine tract-binding protein 2 |
0.322 |
-0.014 |
| 2 |
255333_at |
AT4G04410
|
[copia-like retrotransposon family, has a 1.7e-176 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.316 |
0.021 |
| 3 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
0.204 |
-0.055 |
| 4 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
0.172 |
-0.047 |
| 5 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
0.162 |
-0.132 |
| 6 |
265405_at |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.156 |
-0.182 |
| 7 |
266508_at |
AT2G47920
|
NET3C, Networked 3C |
0.153 |
0.054 |
| 8 |
254043_at |
AT4G25990
|
CIL |
0.149 |
0.211 |
| 9 |
259001_at |
AT3G01960
|
unknown |
0.146 |
-0.090 |
| 10 |
249069_at |
AT5G44010
|
unknown |
0.140 |
-0.147 |
| 11 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.139 |
-0.025 |
| 12 |
249160_at |
AT5G42660
|
unknown |
0.136 |
-0.121 |
| 13 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.134 |
0.216 |
| 14 |
247410_at |
AT5G62990
|
emb1692, embryo defective 1692 |
0.131 |
-0.056 |
| 15 |
264549_at |
AT1G09440
|
[Protein kinase superfamily protein] |
0.131 |
-0.086 |
| 16 |
251001_at |
AT5G02670
|
unknown |
0.126 |
-0.049 |
| 17 |
250722_at |
AT5G06190
|
unknown |
0.123 |
0.002 |
| 18 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
0.119 |
-0.011 |
| 19 |
250379_at |
AT5G11590
|
TINY2, TINY2 |
0.119 |
-0.083 |
| 20 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.117 |
-0.040 |
| 21 |
258509_at |
AT3G06620
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.114 |
-0.001 |
| 22 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.113 |
-0.168 |
| 23 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
0.113 |
0.077 |
| 24 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
0.110 |
-0.144 |
| 25 |
260609_at |
AT2G43690
|
LecRK-V.3, L-type lectin receptor kinase V.3 |
0.107 |
-0.057 |
| 26 |
256080_at |
AT1G20690
|
[SWI-SNF-related chromatin binding protein] |
0.105 |
0.034 |
| 27 |
257837_at |
AT3G25200
|
unknown |
0.105 |
0.001 |
| 28 |
266337_at |
AT2G32390
|
ATGLR3.5, glutamate receptor 3.5, GLR3.5, glutamate receptor 3.5, GLR6 |
0.102 |
-0.012 |
| 29 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.102 |
-0.027 |
| 30 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.100 |
0.137 |
| 31 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.099 |
-0.127 |
| 32 |
253573_at |
AT4G31020
|
[alpha/beta-Hydrolases superfamily protein] |
0.098 |
-0.002 |
| 33 |
263609_at |
AT2G16250
|
[Leucine-rich repeat protein kinase family protein] |
0.097 |
-0.085 |
| 34 |
250587_at |
AT5G07640
|
[RING/U-box superfamily protein] |
0.097 |
-0.050 |
| 35 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
0.096 |
-0.003 |
| 36 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.095 |
-0.049 |
| 37 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.094 |
-0.202 |
| 38 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.093 |
-0.163 |
| 39 |
247518_at |
AT5G61800
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.091 |
-0.042 |
| 40 |
253269_at |
AT4G34140
|
[D111/G-patch domain-containing protein] |
0.091 |
0.099 |
| 41 |
248658_at |
AT5G48600
|
ATCAP-C, ARABIDOPSIS THALIANA CHROMOSOME ASSOCIATED PROTEIN-C, ATSMC3, structural maintenance of chromosome 3, ATSMC4, ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 4, SMC3, structural maintenance of chromosome 3 |
0.090 |
0.041 |
| 42 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.090 |
-0.099 |
| 43 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.088 |
-0.110 |
| 44 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.087 |
-0.040 |
| 45 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
0.087 |
-0.085 |
| 46 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
0.085 |
-0.084 |
| 47 |
249946_at |
AT5G19170
|
unknown |
0.085 |
-0.140 |
| 48 |
250056_at |
AT5G17660
|
[tRNA (guanine-N-7) methyltransferase] |
0.085 |
-0.118 |
| 49 |
246330_at |
AT3G43600
|
AAO2, aldehyde oxidase 2, AO3, aldehyde oxidase 3, AOgamma, Aldehyde oxidase gamma, atAO-2, Arabidopsis thaliana aldehyde oxidase 2, AtAO3, Arabidopsis thaliana aldehyde oxidase 3 |
0.085 |
0.085 |
| 50 |
249468_at |
AT5G39650
|
unknown |
0.084 |
-0.024 |
| 51 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
0.084 |
-0.175 |
| 52 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
0.084 |
-0.132 |
| 53 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-0.412 |
0.535 |
| 54 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
-0.341 |
0.617 |
| 55 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.205 |
0.646 |
| 56 |
255419_at |
AT4G03230
|
[S-locus lectin protein kinase family protein] |
-0.203 |
0.047 |
| 57 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
-0.201 |
0.049 |
| 58 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.194 |
0.135 |
| 59 |
264905_at |
AT2G17430
|
ATMLO7, MLO7, MILDEW RESISTANCE LOCUS O 7, NTA, NORTIA |
-0.190 |
-0.003 |
| 60 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.184 |
0.033 |
| 61 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.184 |
-0.039 |
| 62 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.176 |
0.276 |
| 63 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.170 |
0.224 |
| 64 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.162 |
0.426 |
| 65 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.162 |
0.042 |
| 66 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.162 |
-0.039 |
| 67 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.159 |
-0.163 |
| 68 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
-0.153 |
0.132 |
| 69 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
-0.150 |
0.604 |
| 70 |
259753_at |
AT1G71050
|
HIPP20, heavy metal associated isoprenylated plant protein 20 |
-0.148 |
-0.126 |
| 71 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.148 |
0.054 |
| 72 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.146 |
-0.010 |
| 73 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.145 |
0.676 |
| 74 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.142 |
0.324 |
| 75 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.142 |
-0.104 |
| 76 |
249889_at |
AT5G22540
|
unknown |
-0.141 |
0.159 |
| 77 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.139 |
0.002 |
| 78 |
259401_at |
AT1G17760
|
ATCSTF77, ARABIDOPSIS THALIANA CLEAVAGE STIMULATION FACTOR 77, CSTF77 |
-0.139 |
0.046 |
| 79 |
255806_at |
AT4G10260
|
[pfkB-like carbohydrate kinase family protein] |
-0.139 |
0.018 |
| 80 |
250635_at |
AT5G07510
|
ATGRP-4, ARABIDOPSIS THALIANA GLYCINE-RICH PROTEIN 4, ATGRP14, ARABIDOPSIS THALIANA GLYCINE-RICH PROTEIN 14, GRP-4, GLYCINE-RICH PROTEIN 4, GRP14, glycine-rich protein 14 |
-0.138 |
-0.030 |
| 81 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.136 |
-0.010 |
| 82 |
249262_at |
AT5G41720
|
unknown |
-0.135 |
-0.058 |
| 83 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.133 |
0.558 |
| 84 |
261700_at |
AT1G32690
|
unknown |
-0.131 |
-0.049 |
| 85 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.131 |
-0.057 |
| 86 |
245031_at |
AT2G26360
|
[Mitochondrial substrate carrier family protein] |
-0.131 |
-0.138 |
| 87 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.129 |
-0.047 |
| 88 |
260134_at |
AT1G66370
|
AtMYB113, myb domain protein 113, MYB113, myb domain protein 113 |
-0.126 |
-0.143 |
| 89 |
250400_at |
AT5G10740
|
[Protein phosphatase 2C family protein] |
-0.125 |
-0.036 |
| 90 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.125 |
-0.034 |
| 91 |
253671_at |
AT4G29990
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.125 |
-0.044 |
| 92 |
262989_at |
AT1G23420
|
INO, INNER NO OUTER |
-0.124 |
-0.089 |
| 93 |
248877_at |
AT5G46140
|
unknown |
-0.124 |
-0.093 |
| 94 |
262042_at |
AT1G80140
|
[Pectin lyase-like superfamily protein] |
-0.122 |
-0.082 |
| 95 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
-0.121 |
0.122 |
| 96 |
249747_at |
AT5G24600
|
unknown |
-0.121 |
0.011 |
| 97 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.119 |
0.275 |
| 98 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
-0.118 |
0.040 |
| 99 |
267122_at |
AT2G23550
|
ABE1, ALPHA/BETA FOLD HYDROLASE/ESTERASE 1, ATMES6, METHYL ESTERASE 6, MES6, methyl esterase 6 |
-0.117 |
-0.028 |
| 100 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
-0.117 |
0.014 |
| 101 |
264826_at |
AT1G03410
|
2A6 |
-0.114 |
-0.106 |
| 102 |
256602_at |
AT3G28310
|
unknown |
-0.112 |
-0.093 |
| 103 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
-0.112 |
0.101 |