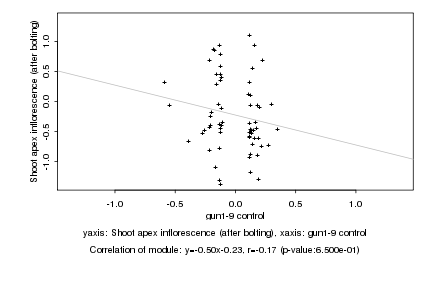
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gun1-9 control |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.343 |
-0.461 |
| 2 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.293 |
-0.048 |
| 3 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.267 |
-0.723 |
| 4 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.218 |
0.689 |
| 5 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.212 |
-0.742 |
| 6 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
0.195 |
-0.086 |
| 7 |
260150_at |
AT1G52820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.191 |
-0.608 |
| 8 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.186 |
-1.298 |
| 9 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.179 |
-0.065 |
| 10 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.178 |
-0.888 |
| 11 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.171 |
-0.436 |
| 12 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.161 |
-0.334 |
| 13 |
248784_at |
AT5G47380
|
unknown |
0.156 |
0.935 |
| 14 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.154 |
-0.612 |
| 15 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.144 |
-0.476 |
| 16 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
0.137 |
0.567 |
| 17 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.135 |
-0.708 |
| 18 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
0.133 |
-0.523 |
| 19 |
267626_at |
AT2G42250
|
CYP712A1, cytochrome P450, family 712, subfamily A, polypeptide 1 |
0.131 |
-0.480 |
| 20 |
264636_at |
AT1G65490
|
unknown |
0.124 |
-0.872 |
| 21 |
253846_at |
AT4G28000
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.122 |
-1.169 |
| 22 |
260599_at |
AT1G55940
|
CYP708A1, cytochrome P450, family 708, subfamily A, polypeptide 1 |
0.121 |
-0.459 |
| 23 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.119 |
-0.051 |
| 24 |
262036_at |
AT1G35530
|
At-FANCM, FANCM, Fanconi anemia complementation group M |
0.119 |
0.101 |
| 25 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.118 |
-0.509 |
| 26 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.116 |
0.319 |
| 27 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.116 |
-0.352 |
| 28 |
251612_at |
AT3G57950
|
unknown |
0.116 |
-0.578 |
| 29 |
250994_at |
AT5G02490
|
AtHsp70-2, Hsp70-2 |
0.116 |
1.104 |
| 30 |
246271_at |
AT4G37230
|
[Photosystem II manganese-stabilising protein (PsbO) family] |
0.111 |
-0.928 |
| 31 |
258271_at |
AT3G15605
|
[nucleic acid binding] |
0.109 |
-0.516 |
| 32 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
0.109 |
-0.591 |
| 33 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.108 |
0.128 |
| 34 |
263242_at |
AT2G31400
|
GUN1, genomes uncoupled 1 |
-0.596 |
0.318 |
| 35 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.548 |
-0.060 |
| 36 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.395 |
-0.655 |
| 37 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.277 |
-0.529 |
| 38 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.264 |
-0.480 |
| 39 |
257670_at |
AT3G20340
|
unknown |
-0.220 |
-0.420 |
| 40 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.218 |
-0.800 |
| 41 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.217 |
0.686 |
| 42 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.214 |
-0.385 |
| 43 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.210 |
-0.243 |
| 44 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.202 |
-0.171 |
| 45 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.189 |
0.870 |
| 46 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.178 |
0.867 |
| 47 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.171 |
-1.097 |
| 48 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.164 |
0.455 |
| 49 |
255711_at |
AT4G00090
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.159 |
0.297 |
| 50 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.147 |
-0.035 |
| 51 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.140 |
-0.380 |
| 52 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.139 |
-1.311 |
| 53 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.138 |
0.940 |
| 54 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.135 |
-0.775 |
| 55 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.131 |
0.458 |
| 56 |
256139_at |
AT1G48660
|
[Auxin-responsive GH3 family protein] |
-0.130 |
-0.443 |
| 57 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.130 |
0.784 |
| 58 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.130 |
0.597 |
| 59 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.127 |
-0.509 |
| 60 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.126 |
0.361 |
| 61 |
255315_at |
AT4G04165
|
[pseudogene] |
-0.126 |
-1.367 |
| 62 |
259508_at |
AT1G43920
|
unknown |
-0.120 |
-0.399 |
| 63 |
248502_at |
AT5G50450
|
[HCP-like superfamily protein with MYND-type zinc finger] |
-0.118 |
-0.115 |
| 64 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.116 |
0.411 |
| 65 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
-0.114 |
-0.335 |