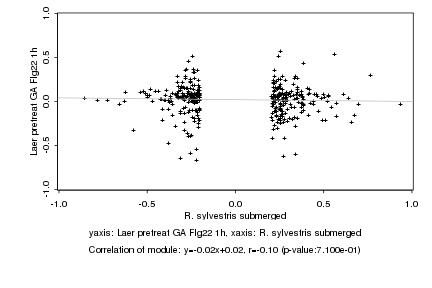
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
R. sylvestris submerged |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.933 |
-0.026 |
| 2 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.760 |
0.299 |
| 3 |
245226_at |
AT3G29970
|
[B12D protein] |
0.697 |
-0.030 |
| 4 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.669 |
-0.159 |
| 5 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
0.654 |
-0.232 |
| 6 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
0.638 |
0.039 |
| 7 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.608 |
0.084 |
| 8 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.571 |
-0.022 |
| 9 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.569 |
-0.161 |
| 10 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
0.561 |
0.540 |
| 11 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.544 |
-0.063 |
| 12 |
265481_at |
AT2G15960
|
unknown |
0.526 |
0.072 |
| 13 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.524 |
0.062 |
| 14 |
263919_at |
AT2G36470
|
unknown |
0.516 |
0.006 |
| 15 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.510 |
-0.213 |
| 16 |
264820_at |
AT1G03475
|
ATCPO-I, HEMF1, LIN2, LESION INITIATION 2 |
0.499 |
0.049 |
| 17 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.494 |
0.086 |
| 18 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.489 |
-0.208 |
| 19 |
249633_at |
AT5G37180
|
ATSUS5, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 5, SUS5, sucrose synthase 5 |
0.487 |
0.043 |
| 20 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.467 |
-0.108 |
| 21 |
260214_at |
AT1G74510
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.460 |
0.068 |
| 22 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
0.458 |
0.088 |
| 23 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.445 |
0.084 |
| 24 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.440 |
0.005 |
| 25 |
253530_at |
AT4G31530
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.440 |
-0.025 |
| 26 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.425 |
-0.017 |
| 27 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.417 |
0.144 |
| 28 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.416 |
0.100 |
| 29 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.410 |
0.082 |
| 30 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.409 |
-0.150 |
| 31 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.404 |
0.151 |
| 32 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.383 |
0.034 |
| 33 |
248302_at |
AT5G53160
|
PYL8, PYR1-like 8, RCAR3, regulatory components of ABA receptor 3 |
0.383 |
-0.030 |
| 34 |
256891_at |
AT3G19030
|
unknown |
0.383 |
-0.100 |
| 35 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.382 |
-0.040 |
| 36 |
250640_at |
AT5G07150
|
[Leucine-rich repeat protein kinase family protein] |
0.380 |
0.442 |
| 37 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.378 |
0.065 |
| 38 |
256081_at |
AT1G20700
|
ATWOX14, WUSCHEL RELATED HOMEOBOX 14, WOX14, WUSCHEL related homeobox 14 |
0.377 |
0.051 |
| 39 |
255883_at |
AT1G20270
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.375 |
-0.012 |
| 40 |
250246_at |
AT5G13700
|
APAO, ATPAO1, polyamine oxidase 1, PAO1, polyamine oxidase 1 |
0.373 |
0.088 |
| 41 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.372 |
-0.059 |
| 42 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.372 |
0.121 |
| 43 |
247320_at |
AT5G64040
|
PSAN |
0.371 |
-0.117 |
| 44 |
253093_at |
AT4G37460
|
SRFR1, SUPPRESSOR OF RPS4-RLD 1 |
0.369 |
-0.012 |
| 45 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.362 |
0.070 |
| 46 |
246335_at |
AT3G44880
|
ACD1, ACCELERATED CELL DEATH 1, LLS1, LETHAL LEAF-SPOT 1 HOMOLOG, PAO, PHEOPHORBIDE A OXYGENASE |
0.355 |
-0.009 |
| 47 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.351 |
0.113 |
| 48 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
0.350 |
0.140 |
| 49 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.350 |
0.164 |
| 50 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
0.349 |
0.040 |
| 51 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.348 |
0.272 |
| 52 |
255435_at |
AT4G03280
|
PETC, photosynthetic electron transfer C, PGR1, PROTON GRADIENT REGULATION 1 |
0.347 |
-0.182 |
| 53 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.345 |
0.090 |
| 54 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.345 |
0.040 |
| 55 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
0.340 |
-0.274 |
| 56 |
258047_at |
AT3G21240
|
4CL2, 4-coumarate:CoA ligase 2, AT4CL2 |
0.340 |
0.294 |
| 57 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.336 |
-0.595 |
| 58 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.333 |
-0.064 |
| 59 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.332 |
0.262 |
| 60 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.332 |
0.033 |
| 61 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
0.331 |
-0.173 |
| 62 |
263305_at |
AT2G01930
|
ATBPC1, BASIC PENTACYSTEINE1, BBR, BPC1, basic pentacysteine1 |
0.327 |
0.078 |
| 63 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.325 |
0.197 |
| 64 |
259017_at |
AT3G07310
|
unknown |
0.318 |
-0.202 |
| 65 |
261669_at |
AT1G18490
|
unknown |
0.317 |
-0.061 |
| 66 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.312 |
0.125 |
| 67 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.311 |
-0.026 |
| 68 |
253472_at |
AT4G32230
|
unknown |
0.308 |
0.061 |
| 69 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.303 |
0.093 |
| 70 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.302 |
-0.183 |
| 71 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.296 |
0.083 |
| 72 |
249384_at |
AT5G39890
|
unknown |
0.296 |
0.082 |
| 73 |
253163_at |
AT4G35750
|
[SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein] |
0.294 |
0.020 |
| 74 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.291 |
-0.085 |
| 75 |
260413_at |
AT1G69800
|
[Cystathionine beta-synthase (CBS) protein] |
0.290 |
-0.225 |
| 76 |
250737_at |
AT5G06370
|
[NC domain-containing protein-related] |
0.287 |
0.014 |
| 77 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.284 |
0.110 |
| 78 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.284 |
0.047 |
| 79 |
248834_at |
AT5G47090
|
unknown |
0.282 |
0.075 |
| 80 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.281 |
0.244 |
| 81 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
0.281 |
-0.130 |
| 82 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.280 |
0.039 |
| 83 |
263804_at |
AT2G40270
|
[Protein kinase family protein] |
0.280 |
0.075 |
| 84 |
263805_at |
AT2G40400
|
unknown |
0.278 |
-0.013 |
| 85 |
259896_at |
AT1G71500
|
[Rieske (2Fe-2S) domain-containing protein] |
0.277 |
-0.198 |
| 86 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.276 |
-0.413 |
| 87 |
248078_at |
AT5G55780
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.273 |
0.090 |
| 88 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.272 |
0.038 |
| 89 |
261919_at |
AT1G65980
|
TPX1, thioredoxin-dependent peroxidase 1 |
0.272 |
0.006 |
| 90 |
258262_at |
AT3G15770
|
unknown |
0.271 |
0.043 |
| 91 |
265843_at |
AT2G35690
|
ACX5, acyl-CoA oxidase 5 |
0.270 |
-0.042 |
| 92 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
0.270 |
-0.621 |
| 93 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.269 |
0.050 |
| 94 |
255365_at |
AT4G04040
|
MEE51, maternal effect embryo arrest 51 |
0.267 |
-0.235 |
| 95 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.265 |
0.034 |
| 96 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
0.265 |
-0.141 |
| 97 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.265 |
-0.167 |
| 98 |
250152_at |
AT5G15120
|
unknown |
0.263 |
0.287 |
| 99 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.262 |
0.140 |
| 100 |
245418_at |
AT4G17370
|
[Oxidoreductase family protein] |
0.262 |
-0.107 |
| 101 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.262 |
0.099 |
| 102 |
253615_at |
AT4G30360
|
ATCNGC17, CYCLIC NUCLEOTIDE-GATED CHANNEL 17, CNGC17, cyclic nucleotide-gated channel 17 |
0.261 |
0.094 |
| 103 |
259967_at |
AT1G76510
|
[ARID/BRIGHT DNA-binding domain-containing protein] |
0.257 |
0.087 |
| 104 |
261003_at |
AT1G26500
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.257 |
-0.137 |
| 105 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.255 |
0.579 |
| 106 |
264673_at |
AT1G09795
|
ATATP-PRT2, ATP phosphoribosyl transferase 2, ATP-PRT2, ATP phosphoribosyl transferase 2, HISN1B |
0.255 |
-0.048 |
| 107 |
256413_at |
AT3G11100
|
[sequence-specific DNA binding transcription factors] |
0.254 |
0.151 |
| 108 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.254 |
-0.084 |
| 109 |
261298_at |
AT1G48510
|
[Surfeit locus 1 cytochrome c oxidase biogenesis protein] |
0.252 |
0.073 |
| 110 |
259528_at |
AT1G12330
|
unknown |
0.252 |
-0.206 |
| 111 |
252027_at |
AT3G52850
|
ATELP, ARABIDOPSIS THALIANA EPIDERMAL GROWTH FACTOR RECEPTOR-LIKE PROTEIN, ATELP1, ATVSR1, BP-80, BP80, BP80-1;1, binding protein of 80 kDa 1;1, BP80B, GFS1, Green fluorescent seed 1, VSR1, vacuolar sorting receptor homolog 1, VSR1;1, VACUOLAR SORTING RECEPTOR 1;1 |
0.252 |
-0.012 |
| 112 |
246184_at |
AT5G20950
|
[Glycosyl hydrolase family protein] |
0.252 |
-0.092 |
| 113 |
248245_at |
AT5G53190
|
AtSWEET3, SWEET3 |
0.252 |
0.080 |
| 114 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.252 |
0.086 |
| 115 |
255258_at |
AT4G05060
|
[PapD-like superfamily protein] |
0.252 |
0.197 |
| 116 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
0.252 |
-0.187 |
| 117 |
265966_at |
AT2G37220
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.251 |
-0.190 |
| 118 |
258455_at |
AT3G22440
|
[FRIGIDA-like protein] |
0.251 |
-0.028 |
| 119 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.251 |
0.256 |
| 120 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
0.251 |
-0.077 |
| 121 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
0.251 |
-0.248 |
| 122 |
266018_at |
AT2G18710
|
SCY1, SECY homolog 1 |
0.251 |
-0.161 |
| 123 |
252949_at |
AT4G38670
|
[Pathogenesis-related thaumatin superfamily protein] |
0.249 |
-0.081 |
| 124 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
0.249 |
0.102 |
| 125 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.248 |
0.177 |
| 126 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.247 |
0.059 |
| 127 |
265943_at |
AT2G19570
|
AT-CDA1, CDA1, cytidine deaminase 1, DESZ |
0.243 |
0.025 |
| 128 |
259863_at |
AT1G72630
|
ELF4-L2, ELF4-like 2 |
0.243 |
-0.195 |
| 129 |
260193_at |
AT1G67640
|
[Transmembrane amino acid transporter family protein] |
0.242 |
0.149 |
| 130 |
262932_at |
AT1G65820
|
[microsomal glutathione s-transferase, putative] |
0.242 |
0.075 |
| 131 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
0.242 |
0.516 |
| 132 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.241 |
0.085 |
| 133 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.240 |
0.254 |
| 134 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.240 |
0.073 |
| 135 |
264981_at |
AT1G27160
|
[valyl-tRNA synthetase / valine--tRNA ligase-related] |
0.240 |
0.023 |
| 136 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
0.237 |
-0.301 |
| 137 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
0.236 |
0.052 |
| 138 |
267214_at |
AT2G43970
|
AtLARP6b, LARP6b, La related protein 6b |
0.236 |
0.025 |
| 139 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.235 |
-0.234 |
| 140 |
263556_at |
AT2G16365
|
[F-box family protein] |
0.235 |
0.150 |
| 141 |
259517_at |
AT1G20630
|
CAT1, catalase 1 |
0.233 |
0.137 |
| 142 |
248827_at |
AT5G47100
|
ATCBL9, CBL9, calcineurin B-like protein 9 |
0.231 |
0.231 |
| 143 |
263647_at |
AT2G04690
|
Pyridoxamine 5'-phosphate oxidase family protein |
0.230 |
-0.152 |
| 144 |
265594_at |
AT2G20060
|
[Ribosomal protein L4/L1 family] |
0.229 |
-0.061 |
| 145 |
260229_at |
AT1G74370
|
[RING/U-box superfamily protein] |
0.228 |
-0.170 |
| 146 |
261757_at |
AT1G08210
|
[Eukaryotic aspartyl protease family protein] |
0.227 |
-0.103 |
| 147 |
266582_at |
AT2G46090
|
LCBK2, long-chain base (LCB) kinase 2 |
0.226 |
-0.077 |
| 148 |
253805_at |
AT4G28260
|
unknown |
0.226 |
0.059 |
| 149 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
0.224 |
-0.113 |
| 150 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
0.224 |
0.046 |
| 151 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
0.223 |
0.023 |
| 152 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.223 |
0.166 |
| 153 |
252293_at |
AT3G48990
|
AAE3, ACYL-ACTIVATING ENZYME 3 |
0.222 |
-0.003 |
| 154 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.221 |
-0.270 |
| 155 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.220 |
0.119 |
| 156 |
248685_at |
AT5G48500
|
unknown |
0.220 |
0.215 |
| 157 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.220 |
0.059 |
| 158 |
250831_at |
AT5G04920
|
[EAP30/Vps36 family protein] |
0.218 |
0.121 |
| 159 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.218 |
0.355 |
| 160 |
261668_at |
AT1G18500
|
IPMS1, ISOPROPYLMALATE SYNTHASE 1, MAML-4, methylthioalkylmalate synthase-like 4 |
0.217 |
0.061 |
| 161 |
253546_at |
AT4G31040
|
[CemA-like proton extrusion protein-related] |
0.217 |
-0.104 |
| 162 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
0.216 |
0.127 |
| 163 |
259969_at |
AT1G76550
|
[Phosphofructokinase family protein] |
0.216 |
0.287 |
| 164 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.215 |
0.247 |
| 165 |
267490_at |
AT2G19130
|
[S-locus lectin protein kinase family protein] |
0.214 |
0.208 |
| 166 |
263799_at |
AT2G24550
|
unknown |
0.214 |
0.052 |
| 167 |
247851_at |
AT5G58070
|
ATTIL, TEMPERATURE-INDUCED LIPOCALIN, TIL, temperature-induced lipocalin |
0.213 |
-0.032 |
| 168 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
0.213 |
0.085 |
| 169 |
259660_at |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
0.212 |
0.011 |
| 170 |
245198_at |
AT1G67700
|
HHL1, HYPERSENSITIVE TO HIGH LIGHT 1 |
0.212 |
-0.309 |
| 171 |
253041_at |
AT4G37870
|
PCK1, phosphoenolpyruvate carboxykinase 1, PEPCK, PHOSPHOENOLPYRUVATE CARBOXYKINASE |
0.212 |
0.164 |
| 172 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.211 |
0.225 |
| 173 |
252049_at |
AT3G52510
|
[F-box associated ubiquitination effector family protein] |
0.210 |
0.029 |
| 174 |
259586_at |
AT1G28100
|
unknown |
0.209 |
-0.050 |
| 175 |
247903_at |
AT5G57340
|
unknown |
0.208 |
-0.131 |
| 176 |
263787_at |
AT2G46420
|
unknown |
0.207 |
-0.412 |
| 177 |
261558_at |
AT1G01770
|
unknown |
0.205 |
0.045 |
| 178 |
257831_at |
AT3G26710
|
CCB1, cofactor assembly of complex C |
0.205 |
-0.205 |
| 179 |
264675_at |
AT1G09830
|
PUR2, purine biosynthesis 2 |
0.205 |
-0.013 |
| 180 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
0.204 |
-0.174 |
| 181 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.203 |
0.067 |
| 182 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.859 |
0.039 |
| 183 |
252640_at |
AT3G44560
|
FAR8, fatty acid reductase 8 |
-0.786 |
0.023 |
| 184 |
250165_at |
AT5G15290
|
CASP5, Casparian strip membrane domain protein 5 |
-0.730 |
0.015 |
| 185 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
-0.661 |
-0.027 |
| 186 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.633 |
0.001 |
| 187 |
263932_at |
AT2G35990
|
LOG2, LONELY GUY 2 |
-0.628 |
0.113 |
| 188 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.581 |
-0.324 |
| 189 |
247722_at |
AT5G59150
|
ATRAB-A2D, ARABIDOPSIS RAB GTPASE HOMOLOG A2D, ATRABA2D, RAB GTPase homolog A2D, RABA2D, RAB GTPase homolog A2D |
-0.540 |
0.112 |
| 190 |
264391_at |
AT1G11920
|
[Pectin lyase-like superfamily protein] |
-0.524 |
0.122 |
| 191 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
-0.510 |
0.100 |
| 192 |
261460_at |
AT1G07880
|
ATMPK13 |
-0.502 |
0.078 |
| 193 |
259064_at |
AT3G07490
|
AGD11, ARF-GAP domain 11, AtCML3, CML3, calmodulin-like 3 |
-0.501 |
0.046 |
| 194 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
-0.500 |
0.077 |
| 195 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.489 |
0.070 |
| 196 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
-0.485 |
0.141 |
| 197 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
-0.472 |
0.008 |
| 198 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.458 |
0.119 |
| 199 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.437 |
0.115 |
| 200 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.427 |
0.029 |
| 201 |
254379_at |
AT4G21820
|
[binding] |
-0.421 |
0.030 |
| 202 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.416 |
-0.087 |
| 203 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.414 |
-0.211 |
| 204 |
262664_at |
AT1G13970
|
unknown |
-0.407 |
0.014 |
| 205 |
263544_at |
AT2G21590
|
APL4 |
-0.401 |
0.018 |
| 206 |
256447_at |
AT1G33440
|
AtNPF4.4, NPF4.4, NRT1/ PTR family 4.4 |
-0.401 |
0.074 |
| 207 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.394 |
0.129 |
| 208 |
252001_at |
AT3G52750
|
FTSZ2-2 |
-0.384 |
-0.476 |
| 209 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.380 |
0.019 |
| 210 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.380 |
-0.083 |
| 211 |
252163_at |
AT3G50610
|
unknown |
-0.377 |
-0.007 |
| 212 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.370 |
0.068 |
| 213 |
249449_at |
AT5G39430
|
unknown |
-0.368 |
0.040 |
| 214 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.364 |
0.007 |
| 215 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
-0.362 |
-0.033 |
| 216 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
-0.362 |
-0.149 |
| 217 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.361 |
0.099 |
| 218 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.340 |
0.085 |
| 219 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
-0.340 |
-0.274 |
| 220 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.334 |
0.073 |
| 221 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.332 |
0.295 |
| 222 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.331 |
0.220 |
| 223 |
247634_at |
AT5G60520
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.330 |
0.098 |
| 224 |
254972_at |
AT4G10440
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.329 |
0.054 |
| 225 |
254400_at |
AT4G21270
|
ATK1, kinesin 1, KATA, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A, KATAP, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA A PROTEIN |
-0.329 |
0.068 |
| 226 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.326 |
0.176 |
| 227 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.326 |
0.066 |
| 228 |
247519_at |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
-0.325 |
0.125 |
| 229 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.323 |
0.041 |
| 230 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
-0.314 |
0.052 |
| 231 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.314 |
0.126 |
| 232 |
252888_at |
AT4G39210
|
APL3 |
-0.313 |
-0.110 |
| 233 |
252115_at |
AT3G51600
|
LTP5, lipid transfer protein 5 |
-0.313 |
-0.647 |
| 234 |
247671_at |
AT5G60210
|
RIP5, ROP interactive partner 5 |
-0.313 |
-0.132 |
| 235 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.312 |
0.149 |
| 236 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.309 |
0.056 |
| 237 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.307 |
0.171 |
| 238 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.305 |
0.064 |
| 239 |
250211_at |
AT5G13880
|
unknown |
-0.303 |
0.221 |
| 240 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.302 |
0.094 |
| 241 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.299 |
0.064 |
| 242 |
254958_at |
AT4G11010
|
NDPK3, nucleoside diphosphate kinase 3 |
-0.299 |
0.030 |
| 243 |
264188_at |
AT1G54690
|
G-H2AX, GAMMA H2AX, GAMMA-H2AX, gamma histone variant H2AX, H2AXB, HTA3, histone H2A 3 |
-0.297 |
-0.076 |
| 244 |
257814_at |
AT3G25110
|
AtFaTA, fatA acyl-ACP thioesterase, FaTA, fatA acyl-ACP thioesterase |
-0.296 |
0.037 |
| 245 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.296 |
0.162 |
| 246 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.295 |
0.060 |
| 247 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
-0.294 |
-0.230 |
| 248 |
254374_at |
AT4G21780
|
unknown |
-0.294 |
0.020 |
| 249 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.294 |
0.072 |
| 250 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.294 |
0.150 |
| 251 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.293 |
-0.326 |
| 252 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.293 |
-0.130 |
| 253 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.291 |
0.076 |
| 254 |
246185_at |
AT5G20980
|
ATMS3, methionine synthase 3, MS3, methionine synthase 3 |
-0.290 |
0.152 |
| 255 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.288 |
0.358 |
| 256 |
245772_at |
AT1G30300
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.286 |
0.263 |
| 257 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.285 |
0.285 |
| 258 |
263701_at |
AT1G31160
|
HINT 2, HISTIDINE TRIAD NUCLEOTIDE-BINDING 2 |
-0.284 |
-0.061 |
| 259 |
247039_at |
AT5G67270
|
ATEB1C, MICROTUBULE END BINDING PROTEIN 1, ATEB1H1, ATEB1-HOMOLOG1, EB1C, end binding protein 1C |
-0.281 |
0.052 |
| 260 |
263971_at |
AT2G42800
|
AtRLP29, receptor like protein 29, RLP29, receptor like protein 29 |
-0.280 |
0.150 |
| 261 |
248139_at |
AT5G54970
|
unknown |
-0.278 |
0.369 |
| 262 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.276 |
-0.356 |
| 263 |
267574_at |
AT2G30680
|
unknown |
-0.274 |
0.095 |
| 264 |
260636_at |
AT1G62430
|
ATCDS1, CDP-diacylglycerol synthase 1, CDS1, CDP-diacylglycerol synthase 1 |
-0.273 |
0.152 |
| 265 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.272 |
0.064 |
| 266 |
261199_at |
AT1G12950
|
RSH2, root hair specific 2 |
-0.272 |
0.065 |
| 267 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.271 |
0.459 |
| 268 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.271 |
0.119 |
| 269 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.270 |
-0.391 |
| 270 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
-0.270 |
-0.122 |
| 271 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
-0.269 |
-0.011 |
| 272 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.265 |
0.083 |
| 273 |
259095_at |
AT3G05020
|
ACP, ACYL CARRIER PROTEIN, ACP1, acyl carrier protein 1 |
-0.262 |
-0.098 |
| 274 |
247425_at |
AT5G62550
|
unknown |
-0.262 |
0.027 |
| 275 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
-0.260 |
0.257 |
| 276 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.259 |
0.230 |
| 277 |
249131_at |
AT5G43110
|
APUM14, pumilio 14, PUM14, pumilio 14 |
-0.259 |
0.091 |
| 278 |
256575_at |
AT3G14790
|
ATRHM3, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 3, RHM3, rhamnose biosynthesis 3 |
-0.259 |
0.149 |
| 279 |
267434_at |
AT2G26260
|
3BETAHSD/D2, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2, AT3BETAHSD/D2, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 |
-0.258 |
0.040 |
| 280 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
-0.257 |
0.074 |
| 281 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.257 |
-0.585 |
| 282 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.257 |
-0.389 |
| 283 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.255 |
0.225 |
| 284 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.253 |
0.086 |
| 285 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
-0.253 |
0.094 |
| 286 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.253 |
0.088 |
| 287 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.251 |
0.002 |
| 288 |
265545_at |
AT2G28250
|
NCRK |
-0.250 |
0.083 |
| 289 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.249 |
-0.386 |
| 290 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.248 |
0.065 |
| 291 |
245301_at |
AT4G17190
|
FPS2, farnesyl diphosphate synthase 2 |
-0.248 |
0.090 |
| 292 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
-0.247 |
0.191 |
| 293 |
257931_at |
AT3G17030
|
[Nucleic acid-binding proteins superfamily] |
-0.247 |
0.045 |
| 294 |
252973_s_at |
AT4G38740
|
ROC1, rotamase CYP 1 |
-0.246 |
-0.093 |
| 295 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
-0.246 |
0.049 |
| 296 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
-0.245 |
0.521 |
| 297 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.245 |
-0.172 |
| 298 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.243 |
0.014 |
| 299 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.242 |
0.365 |
| 300 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.241 |
0.337 |
| 301 |
262802_at |
AT1G20930
|
CDKB2;2, cyclin-dependent kinase B2;2 |
-0.241 |
0.141 |
| 302 |
252335_at |
AT3G48860
|
SCD2, STOMATAL CYTOKINESIS DEFECTIVE 2 |
-0.241 |
0.077 |
| 303 |
267308_at |
AT2G30200
|
EMB3147, EMBRYO DEFECTIVE 3147 |
-0.240 |
-0.001 |
| 304 |
247225_at |
AT5G65090
|
BST1, BRISTLED 1, DER4, DEFORMED ROOT HAIRS 4, MRH3 |
-0.238 |
0.018 |
| 305 |
262686_at |
AT1G75990
|
[PAM domain (PCI/PINT associated module) protein] |
-0.237 |
0.187 |
| 306 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
-0.237 |
0.003 |
| 307 |
253486_at |
AT4G31600
|
UTr7, UDP-galactose transporter 7 |
-0.237 |
-0.015 |
| 308 |
250778_at |
AT5G05500
|
MOP10 |
-0.236 |
0.052 |
| 309 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.236 |
-0.220 |
| 310 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
-0.236 |
0.099 |
| 311 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.236 |
0.269 |
| 312 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.234 |
0.053 |
| 313 |
256160_at |
AT1G30120
|
PDH-E1 BETA, pyruvate dehydrogenase E1 beta |
-0.233 |
0.002 |
| 314 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.233 |
0.049 |
| 315 |
248146_at |
AT5G54940
|
[Translation initiation factor SUI1 family protein] |
-0.232 |
0.028 |
| 316 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.232 |
-0.193 |
| 317 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.232 |
0.333 |
| 318 |
260643_at |
AT1G53270
|
ABCG10, ATP-binding cassette G10 |
-0.231 |
0.042 |
| 319 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.229 |
-0.093 |
| 320 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.227 |
-0.004 |
| 321 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.225 |
-0.009 |
| 322 |
267367_at |
AT2G44210
|
unknown |
-0.224 |
-0.102 |
| 323 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.222 |
-0.665 |
| 324 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.222 |
0.073 |
| 325 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.221 |
-0.537 |
| 326 |
249473_at |
AT5G39340
|
AHP3, histidine-containing phosphotransmitter 3, ATHP2, ARABIDOPSIS THALIANA HISTIDINE-CONTAINING PHOSPHOTRANSMITTER 2 |
-0.221 |
0.083 |
| 327 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
-0.220 |
0.363 |
| 328 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.220 |
0.077 |
| 329 |
257087_at |
AT3G20500
|
ATPAP18, PURPLE ACID PHOSPHATASE 18, PAP18, purple acid phosphatase 18 |
-0.220 |
-0.001 |
| 330 |
251924_at |
AT3G53730
|
[Histone superfamily protein] |
-0.219 |
0.099 |
| 331 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.219 |
-0.002 |
| 332 |
267582_at |
AT2G41970
|
[Protein kinase superfamily protein] |
-0.219 |
0.016 |
| 333 |
256964_at |
AT3G13520
|
AGP12, arabinogalactan protein 12, ATAGP12 |
-0.217 |
0.103 |
| 334 |
246505_at |
AT5G16250
|
unknown |
-0.217 |
0.126 |
| 335 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.215 |
-0.159 |
| 336 |
262811_at |
AT1G11700
|
unknown |
-0.215 |
-0.121 |
| 337 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.214 |
-0.244 |
| 338 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.214 |
0.128 |
| 339 |
264565_at |
AT1G05280
|
unknown |
-0.214 |
0.185 |
| 340 |
263552_x_at |
AT2G24980
|
EXT6, extensin 6 |
-0.214 |
0.135 |
| 341 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.213 |
0.245 |
| 342 |
249946_at |
AT5G19170
|
unknown |
-0.211 |
0.077 |
| 343 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.211 |
0.062 |
| 344 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.210 |
-0.188 |
| 345 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
-0.210 |
-0.293 |
| 346 |
259339_at |
AT3G03900
|
adenosine-5'-phosphosulfate (APS) kinase 3 |
-0.209 |
0.100 |
| 347 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
-0.209 |
0.134 |
| 348 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.208 |
0.032 |
| 349 |
247746_at |
AT5G58970
|
ATUCP2, uncoupling protein 2, UCP2, uncoupling protein 2 |
-0.207 |
-0.209 |
| 350 |
258663_at |
AT3G08670
|
unknown |
-0.207 |
-0.096 |
| 351 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.207 |
0.054 |
| 352 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.206 |
-0.070 |
| 353 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
-0.206 |
0.018 |
| 354 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.206 |
0.056 |
| 355 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
-0.206 |
0.078 |
| 356 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.205 |
0.073 |
| 357 |
259757_at |
AT1G77510
|
ATPDI6, PROTEIN DISULFIDE ISOMERASE 6, ATPDIL1-2, PDI-like 1-2, PDI6, PROTEIN DISULFIDE ISOMERASE 6, PDIL1-2, PDI-like 1-2 |
-0.205 |
0.163 |
| 358 |
261754_at |
AT1G76130
|
AMY2, alpha-amylase-like 2, ATAMY2, ARABIDOPSIS THALIANA ALPHA-AMYLASE-LIKE 2 |
-0.205 |
-0.081 |
| 359 |
265243_at |
AT2G43040
|
NPG1, no pollen germination 1 |
-0.205 |
0.099 |
| 360 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
-0.204 |
0.056 |
| 361 |
250386_at |
AT5G11510
|
AtMYB3R4, myb domain protein 3R4, MYB3R-4, myb domain protein 3r-4 |
-0.204 |
0.091 |