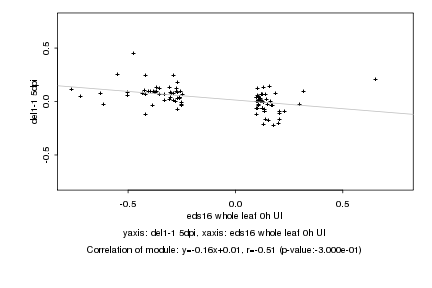
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
eds16 whole leaf 0h UI |
Log2 signal ratio
del1-1 5dpi |
| 1 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.650 |
0.208 |
| 2 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.313 |
0.095 |
| 3 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.298 |
-0.026 |
| 4 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
0.225 |
-0.088 |
| 5 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.204 |
-0.164 |
| 6 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.202 |
-0.092 |
| 7 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.202 |
-0.105 |
| 8 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.196 |
-0.198 |
| 9 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.182 |
0.080 |
| 10 |
264343_at |
AT1G11850
|
unknown |
0.173 |
-0.218 |
| 11 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
0.168 |
-0.033 |
| 12 |
254349_at |
AT4G22250
|
[RING/U-box superfamily protein] |
0.167 |
-0.029 |
| 13 |
260506_at |
AT1G47210
|
CYCA3;2, cyclin-dependent protein kinase 3;2 |
0.163 |
0.003 |
| 14 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
0.154 |
0.141 |
| 15 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.151 |
-0.173 |
| 16 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
0.149 |
-0.022 |
| 17 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.143 |
0.020 |
| 18 |
248091_at |
AT5G55120
|
galactose-1-phosphate guanylyltransferase (GDP)s;GDP-D-glucose phosphorylases;quercetin 4'-O-glucosyltransferases |
0.140 |
0.073 |
| 19 |
251591_at |
AT3G57680
|
[Peptidase S41 family protein] |
0.136 |
-0.167 |
| 20 |
250642_at |
AT5G07180
|
ERL2, ERECTA-like 2 |
0.135 |
-0.086 |
| 21 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
0.134 |
-0.066 |
| 22 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.131 |
-0.087 |
| 23 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.129 |
-0.003 |
| 24 |
250361_at |
AT5G11370
|
[FBD / Leucine Rich Repeat domains containing protein] |
0.127 |
0.137 |
| 25 |
247399_at |
AT5G62960
|
unknown |
0.127 |
-0.206 |
| 26 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
0.125 |
-0.065 |
| 27 |
260260_at |
AT1G68540
|
CCRL6, cinnamoyl coA reductase-like 6, TKPR2, tetraketide alpha-pyrone reductase 2 |
0.124 |
-0.057 |
| 28 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.123 |
0.005 |
| 29 |
245467_at |
AT4G16610
|
[C2H2-like zinc finger protein] |
0.123 |
-0.065 |
| 30 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.123 |
0.074 |
| 31 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
0.120 |
0.067 |
| 32 |
255460_at |
AT4G02800
|
unknown |
0.118 |
0.006 |
| 33 |
261001_at |
AT1G26530
|
[PIN domain-like family protein] |
0.118 |
0.072 |
| 34 |
264289_at |
AT1G61890
|
[MATE efflux family protein] |
0.115 |
0.036 |
| 35 |
263436_at |
AT2G28690
|
unknown |
0.113 |
0.003 |
| 36 |
254929_at |
AT4G11440
|
[Mitochondrial substrate carrier family protein] |
0.108 |
0.024 |
| 37 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.107 |
-0.025 |
| 38 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
0.105 |
0.045 |
| 39 |
259005_at |
AT3G01930
|
[Major facilitator superfamily protein] |
0.105 |
0.015 |
| 40 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.103 |
-0.033 |
| 41 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.103 |
0.048 |
| 42 |
265879_at |
AT2G42450
|
[alpha/beta-Hydrolases superfamily protein] |
0.102 |
0.123 |
| 43 |
258704_at |
AT3G09780
|
ATCRR1, CCR1, CRINKLY4 related 1 |
0.102 |
0.059 |
| 44 |
261181_at |
AT1G34580
|
[Major facilitator superfamily protein] |
0.101 |
0.009 |
| 45 |
252877_at |
AT4G39630
|
unknown |
0.100 |
-0.057 |
| 46 |
252666_at |
AT3G44180
|
[syntaxin-related family protein] |
0.096 |
-0.058 |
| 47 |
250952_at |
AT5G03570
|
ATIREG2, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 2, FPN2, FERROPORTIN 2, IREG2, iron regulated 2 |
0.096 |
0.041 |
| 48 |
246793_at |
AT5G27210
|
Cand8, candidate G-protein Coupled Receptor 8 |
0.095 |
0.040 |
| 49 |
249090_at |
AT5G43745
|
unknown |
0.095 |
-0.121 |
| 50 |
255282_at |
AT4G04600
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.095 |
0.003 |
| 51 |
259385_at |
AT1G13470
|
unknown |
-0.767 |
0.116 |
| 52 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.726 |
0.048 |
| 53 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.630 |
0.082 |
| 54 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.616 |
-0.024 |
| 55 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.551 |
0.261 |
| 56 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.507 |
0.085 |
| 57 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.504 |
0.062 |
| 58 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.476 |
0.454 |
| 59 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.433 |
0.079 |
| 60 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.424 |
0.105 |
| 61 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.423 |
-0.120 |
| 62 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.423 |
0.072 |
| 63 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-0.419 |
0.247 |
| 64 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.406 |
0.102 |
| 65 |
259489_at |
AT1G15790
|
unknown |
-0.400 |
0.102 |
| 66 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.387 |
-0.033 |
| 67 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.382 |
0.095 |
| 68 |
265993_at |
AT2G24160
|
[pseudogene] |
-0.380 |
0.091 |
| 69 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.377 |
0.085 |
| 70 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.372 |
0.100 |
| 71 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.370 |
0.132 |
| 72 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.358 |
0.130 |
| 73 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.354 |
0.075 |
| 74 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.333 |
0.018 |
| 75 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.331 |
0.075 |
| 76 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.311 |
0.133 |
| 77 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.309 |
0.024 |
| 78 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.306 |
0.092 |
| 79 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.305 |
0.042 |
| 80 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
-0.300 |
0.077 |
| 81 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.292 |
0.079 |
| 82 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.291 |
0.011 |
| 83 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
-0.289 |
0.250 |
| 84 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
-0.283 |
0.009 |
| 85 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
-0.277 |
0.124 |
| 86 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.275 |
0.100 |
| 87 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
-0.274 |
0.032 |
| 88 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.274 |
0.187 |
| 89 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
-0.271 |
-0.074 |
| 90 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.270 |
0.089 |
| 91 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.264 |
0.045 |
| 92 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
-0.262 |
0.044 |
| 93 |
250941_at |
AT5G03320
|
[Protein kinase superfamily protein] |
-0.261 |
0.031 |
| 94 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.261 |
0.039 |
| 95 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
-0.260 |
0.095 |
| 96 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
-0.255 |
-0.034 |
| 97 |
246943_at |
AT5G25440
|
[Protein kinase superfamily protein] |
-0.254 |
-0.003 |
| 98 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.253 |
-0.027 |
| 99 |
257101_at |
AT3G25020
|
AtRLP42, receptor like protein 42, RBPG1, response to Botrytis polygalacturonases 1, RLP42, receptor like protein 42 |
-0.248 |
0.069 |