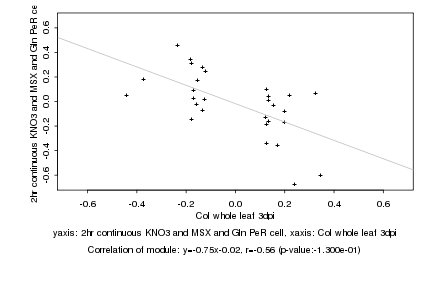
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col whole leaf 3dpi |
Log2 signal ratio
2hr continuous KNO3 and MSX and Gln PeR cell |
| 1 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.345 |
-0.600 |
| 2 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.321 |
0.073 |
| 3 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.238 |
-0.668 |
| 4 |
264774_at |
AT1G22890
|
unknown |
0.217 |
0.050 |
| 5 |
254571_at |
AT4G19370
|
unknown |
0.199 |
-0.080 |
| 6 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.198 |
-0.167 |
| 7 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.167 |
-0.357 |
| 8 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.151 |
-0.028 |
| 9 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
0.134 |
0.043 |
| 10 |
260179_at |
AT1G70690
|
HWI1, HOPW1-1-INDUCED GENE1, PDLP5, PLASMODESMATA-LOCATED PROTEIN 5 |
0.133 |
-0.157 |
| 11 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
0.131 |
0.012 |
| 12 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.125 |
-0.337 |
| 13 |
249839_at |
AT5G23405
|
[HMG-box (high mobility group) DNA-binding family protein] |
0.124 |
0.105 |
| 14 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.123 |
-0.180 |
| 15 |
265128_at |
AT1G30860
|
[RING/U-box superfamily protein] |
0.119 |
-0.127 |
| 16 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.444 |
0.053 |
| 17 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.375 |
0.181 |
| 18 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.238 |
0.463 |
| 19 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.185 |
0.349 |
| 20 |
262236_at |
AT1G48330
|
unknown |
-0.181 |
0.311 |
| 21 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.179 |
-0.144 |
| 22 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.173 |
0.028 |
| 23 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.172 |
0.092 |
| 24 |
256021_at |
AT1G58270
|
ZW9 |
-0.162 |
-0.024 |
| 25 |
255825_at |
AT2G40475
|
ASG8, ALTERED SEED GERMINATION 8 |
-0.154 |
0.176 |
| 26 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.137 |
-0.072 |
| 27 |
261921_at |
AT1G65900
|
unknown |
-0.136 |
0.279 |
| 28 |
261203_at |
AT1G12845
|
unknown |
-0.129 |
0.024 |
| 29 |
261080_at |
AT1G07370
|
ATPCNA1, PROLIFERATING CELLULAR NUCLEAR ANTIGEN 1, PCNA1, proliferating cellular nuclear antigen 1 |
-0.125 |
0.245 |