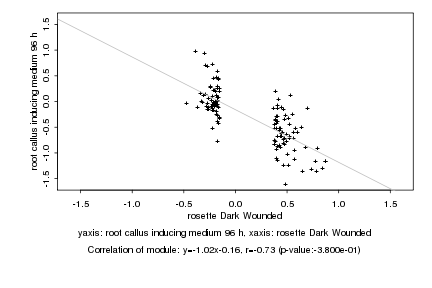
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rosette Dark Wounded |
Log2 signal ratio
root callus inducing medium 96 h |
| 1 |
253832_at |
AT4G27654
|
unknown |
0.864 |
-1.148 |
| 2 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.863 |
-1.166 |
| 3 |
251950_at |
AT3G53600
|
[C2H2-type zinc finger family protein] |
0.843 |
-1.294 |
| 4 |
252739_at |
AT3G43250
|
unknown |
0.792 |
-0.910 |
| 5 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.780 |
-1.342 |
| 6 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.768 |
-1.160 |
| 7 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.728 |
-1.305 |
| 8 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.690 |
-0.124 |
| 9 |
249522_at |
AT5G38700
|
unknown |
0.676 |
-0.879 |
| 10 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.647 |
-1.350 |
| 11 |
245677_at |
AT1G56660
|
unknown |
0.635 |
-0.499 |
| 12 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.593 |
-0.601 |
| 13 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
0.581 |
-0.512 |
| 14 |
253859_at |
AT4G27657
|
unknown |
0.572 |
-1.115 |
| 15 |
265618_at |
AT2G25460
|
unknown |
0.570 |
-0.935 |
| 16 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.561 |
-0.710 |
| 17 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
0.559 |
-0.586 |
| 18 |
256884_at |
AT3G15200
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.544 |
-0.241 |
| 19 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.528 |
0.118 |
| 20 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.522 |
-0.715 |
| 21 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.522 |
-0.670 |
| 22 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.515 |
-0.442 |
| 23 |
248959_at |
AT5G45630
|
unknown |
0.513 |
-0.328 |
| 24 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.506 |
-1.231 |
| 25 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.504 |
-1.019 |
| 26 |
250098_at |
AT5G17350
|
unknown |
0.492 |
-0.761 |
| 27 |
253830_at |
AT4G27652
|
unknown |
0.489 |
-0.631 |
| 28 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.482 |
-0.259 |
| 29 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.477 |
-1.595 |
| 30 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.468 |
-0.827 |
| 31 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.467 |
-0.341 |
| 32 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.466 |
-0.702 |
| 33 |
265632_at |
AT2G14290
|
unknown |
0.463 |
-0.813 |
| 34 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
0.463 |
-0.737 |
| 35 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.462 |
-1.238 |
| 36 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
0.459 |
-0.138 |
| 37 |
253799_at |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
0.451 |
-0.595 |
| 38 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.446 |
-0.108 |
| 39 |
263754_at |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.444 |
-0.667 |
| 40 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.443 |
-0.606 |
| 41 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.436 |
-0.512 |
| 42 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.430 |
-0.672 |
| 43 |
253643_at |
AT4G29780
|
unknown |
0.427 |
-0.886 |
| 44 |
261101_at |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
0.425 |
-0.509 |
| 45 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.420 |
-0.852 |
| 46 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.418 |
-0.550 |
| 47 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.414 |
0.054 |
| 48 |
256044_at |
AT1G07160
|
[Protein phosphatase 2C family protein] |
0.406 |
-0.372 |
| 49 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.406 |
-0.873 |
| 50 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.405 |
-0.668 |
| 51 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.405 |
-0.284 |
| 52 |
262211_at |
AT1G74930
|
ORA47 |
0.405 |
-1.141 |
| 53 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.402 |
-0.135 |
| 54 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.399 |
-0.059 |
| 55 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.397 |
-0.283 |
| 56 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.395 |
-0.920 |
| 57 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.392 |
-0.518 |
| 58 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.391 |
-0.409 |
| 59 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.390 |
-1.093 |
| 60 |
262452_at |
AT1G11210
|
unknown |
0.386 |
-0.763 |
| 61 |
247933_at |
AT5G56980
|
unknown |
0.385 |
-0.339 |
| 62 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.383 |
0.198 |
| 63 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.381 |
-0.365 |
| 64 |
255884_at |
AT1G20310
|
unknown |
0.374 |
-0.753 |
| 65 |
247047_at |
AT5G66650
|
unknown |
0.372 |
-0.818 |
| 66 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.372 |
-0.818 |
| 67 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
0.372 |
-0.435 |
| 68 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.370 |
-0.519 |
| 69 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.365 |
-0.119 |
| 70 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.475 |
-0.028 |
| 71 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.389 |
0.990 |
| 72 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.377 |
-0.101 |
| 73 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.339 |
0.166 |
| 74 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.332 |
0.014 |
| 75 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.320 |
-0.010 |
| 76 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.315 |
0.125 |
| 77 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.303 |
0.946 |
| 78 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.297 |
0.708 |
| 79 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.294 |
0.138 |
| 80 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.282 |
-0.083 |
| 81 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.278 |
-0.145 |
| 82 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.271 |
-0.029 |
| 83 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.271 |
0.684 |
| 84 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.270 |
-0.113 |
| 85 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.267 |
0.078 |
| 86 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.265 |
-0.138 |
| 87 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.251 |
0.280 |
| 88 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.247 |
0.299 |
| 89 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.241 |
-0.094 |
| 90 |
251183_at |
AT3G62630
|
unknown |
-0.232 |
0.033 |
| 91 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.231 |
0.114 |
| 92 |
259373_at |
AT1G69160
|
unknown |
-0.231 |
-0.509 |
| 93 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.229 |
0.737 |
| 94 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.229 |
-0.082 |
| 95 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.228 |
-0.043 |
| 96 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.226 |
-0.131 |
| 97 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.226 |
-0.162 |
| 98 |
265892_at |
AT2G15020
|
unknown |
-0.221 |
-0.159 |
| 99 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.220 |
-0.005 |
| 100 |
247878_at |
AT5G57760
|
unknown |
-0.218 |
-0.161 |
| 101 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.216 |
0.216 |
| 102 |
260081_at |
AT1G78170
|
unknown |
-0.213 |
0.448 |
| 103 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.211 |
-0.023 |
| 104 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.209 |
-0.070 |
| 105 |
260167_at |
AT1G71970
|
unknown |
-0.207 |
0.226 |
| 106 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.207 |
-0.030 |
| 107 |
267535_at |
AT2G41940
|
ZFP8, zinc finger protein 8 |
-0.206 |
-0.096 |
| 108 |
262598_at |
AT1G15260
|
unknown |
-0.204 |
-0.086 |
| 109 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
-0.198 |
0.017 |
| 110 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.197 |
-0.044 |
| 111 |
263688_at |
AT1G26920
|
unknown |
-0.195 |
-0.001 |
| 112 |
254810_at |
AT4G12390
|
PME1, pectin methylesterase inhibitor 1 |
-0.194 |
0.204 |
| 113 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.191 |
-0.251 |
| 114 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
-0.189 |
0.478 |
| 115 |
261538_at |
AT1G01830
|
[ARM repeat superfamily protein] |
-0.188 |
0.119 |
| 116 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.188 |
-0.053 |
| 117 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.184 |
-0.191 |
| 118 |
259103_at |
AT3G11690
|
unknown |
-0.181 |
0.260 |
| 119 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.181 |
0.012 |
| 120 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.180 |
-0.766 |
| 121 |
257610_at |
AT3G13810
|
AtIDD11, indeterminate(ID)-domain 11, IDD11, indeterminate(ID)-domain 11 |
-0.180 |
-0.056 |
| 122 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.179 |
0.310 |
| 123 |
263597_at |
AT2G01870
|
unknown |
-0.178 |
0.020 |
| 124 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
-0.178 |
0.255 |
| 125 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
-0.177 |
-0.093 |
| 126 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.176 |
0.594 |
| 127 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.174 |
0.084 |
| 128 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.174 |
-0.270 |
| 129 |
266110_at |
AT2G02080
|
AtIDD4, indeterminate(ID)-domain 4, IDD4, indeterminate(ID)-domain 4 |
-0.174 |
-0.387 |
| 130 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
-0.173 |
0.081 |
| 131 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.171 |
0.458 |
| 132 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.171 |
-0.412 |
| 133 |
251412_at |
AT3G60220
|
ATL4, TOXICOS EN LEVADURA 4, TL4, TOXICOS EN LEVADURA 4 |
-0.169 |
0.445 |
| 134 |
262711_at |
AT1G16500
|
unknown |
-0.165 |
-0.112 |
| 135 |
245431_at |
AT4G17080
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.163 |
0.203 |
| 136 |
267561_at |
AT2G45590
|
[Protein kinase superfamily protein] |
-0.161 |
0.269 |
| 137 |
252322_at |
AT3G48550
|
unknown |
-0.161 |
-0.326 |
| 138 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.159 |
-0.295 |