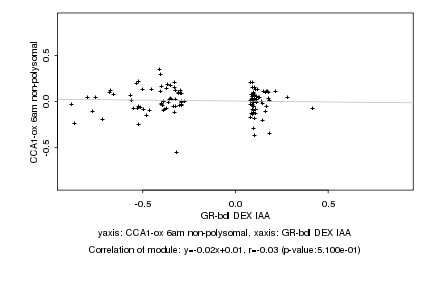
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
GR-bdl DEX IAA |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
0.415 |
-0.074 |
| 2 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
0.277 |
0.051 |
| 3 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.213 |
0.116 |
| 4 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.181 |
-0.346 |
| 5 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
0.179 |
0.013 |
| 6 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
0.176 |
0.108 |
| 7 |
250872_at |
AT5G03960
|
IQD12, IQ-domain 12 |
0.175 |
0.042 |
| 8 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
0.171 |
0.118 |
| 9 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.166 |
0.114 |
| 10 |
254520_at |
AT4G19960
|
ATKUP9, HAK9, KT9, KUP9, K+ uptake permease 9 |
0.163 |
-0.048 |
| 11 |
262811_at |
AT1G11700
|
unknown |
0.159 |
-0.099 |
| 12 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.159 |
0.108 |
| 13 |
253317_at |
AT4G33960
|
unknown |
0.147 |
0.109 |
| 14 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.143 |
-0.019 |
| 15 |
264636_at |
AT1G65490
|
unknown |
0.141 |
-0.202 |
| 16 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.136 |
0.004 |
| 17 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.126 |
0.049 |
| 18 |
263915_at |
AT2G36430
|
unknown |
0.119 |
0.051 |
| 19 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
0.118 |
0.141 |
| 20 |
248140_at |
AT5G54980
|
[Uncharacterised protein family (UPF0497)] |
0.112 |
-0.010 |
| 21 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.111 |
0.027 |
| 22 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.111 |
0.055 |
| 23 |
258156_at |
AT3G18050
|
unknown |
0.109 |
0.027 |
| 24 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
0.109 |
0.011 |
| 25 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.108 |
-0.119 |
| 26 |
254670_at |
AT4G18390
|
TCP2, TCP2, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 |
0.107 |
0.131 |
| 27 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.106 |
-0.079 |
| 28 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.106 |
-0.076 |
| 29 |
246756_at |
AT5G27930
|
[Protein phosphatase 2C family protein] |
0.105 |
-0.077 |
| 30 |
259863_at |
AT1G72630
|
ELF4-L2, ELF4-like 2 |
0.100 |
0.152 |
| 31 |
264865_at |
AT1G24120
|
ARL1, ARG1-like 1 |
0.099 |
-0.367 |
| 32 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.099 |
0.067 |
| 33 |
246971_at |
AT5G24940
|
[Protein phosphatase 2C family protein] |
0.098 |
0.042 |
| 34 |
258959_at |
AT3G10600
|
CAT7, cationic amino acid transporter 7 |
0.098 |
-0.183 |
| 35 |
264115_at |
AT2G31290
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.097 |
0.080 |
| 36 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
0.096 |
-0.052 |
| 37 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.096 |
0.100 |
| 38 |
254777_at |
AT4G12960
|
AtGILT, GILT, gamma-interferon-responsive lysosomal thiol reductase |
0.095 |
-0.115 |
| 39 |
250806_at |
AT5G05070
|
[DHHC-type zinc finger family protein] |
0.094 |
-0.095 |
| 40 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.094 |
0.018 |
| 41 |
262718_at |
AT1G43570
|
unknown |
0.094 |
-0.285 |
| 42 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.093 |
-0.001 |
| 43 |
259476_at |
AT1G19000
|
[Homeodomain-like superfamily protein] |
0.093 |
-0.048 |
| 44 |
261401_at |
AT1G79640
|
[Protein kinase superfamily protein] |
0.092 |
0.093 |
| 45 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.091 |
-0.027 |
| 46 |
256813_at |
AT3G21360
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.090 |
0.059 |
| 47 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
0.090 |
0.157 |
| 48 |
259541_at |
AT1G20650
|
ASG5, ALTERED SEED GERMINATION 5 |
0.089 |
-0.008 |
| 49 |
259989_at |
AT1G41750
|
unknown |
0.089 |
-0.080 |
| 50 |
266086_at |
AT2G38060
|
PHT4;2, phosphate transporter 4;2 |
0.089 |
-0.039 |
| 51 |
249530_at |
AT5G38750
|
[asparaginyl-tRNA synthetase family] |
0.088 |
0.206 |
| 52 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
0.088 |
0.058 |
| 53 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
0.088 |
-0.132 |
| 54 |
248623_at |
AT5G49170
|
unknown |
0.087 |
0.040 |
| 55 |
267426_at |
AT2G34820
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.086 |
0.030 |
| 56 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.083 |
0.080 |
| 57 |
257260_at |
AT3G22104
|
[Phototropic-responsive NPH3 family protein] |
0.082 |
-0.125 |
| 58 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
0.081 |
0.208 |
| 59 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
0.081 |
-0.168 |
| 60 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
0.081 |
0.014 |
| 61 |
255193_at |
AT4G07400
|
VFB3, VIER F-box proteine 3 |
0.081 |
-0.023 |
| 62 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.888 |
-0.032 |
| 63 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-0.869 |
-0.231 |
| 64 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.801 |
0.052 |
| 65 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.773 |
-0.099 |
| 66 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.757 |
0.051 |
| 67 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.721 |
-0.193 |
| 68 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.684 |
0.101 |
| 69 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.678 |
0.123 |
| 70 |
266611_at |
AT2G14960
|
GH3.1 |
-0.659 |
0.083 |
| 71 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.571 |
0.074 |
| 72 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.562 |
0.020 |
| 73 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.550 |
-0.073 |
| 74 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.537 |
0.199 |
| 75 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.529 |
-0.069 |
| 76 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.527 |
0.225 |
| 77 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.524 |
-0.048 |
| 78 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.523 |
-0.240 |
| 79 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.517 |
-0.055 |
| 80 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.502 |
0.135 |
| 81 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.499 |
-0.084 |
| 82 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.482 |
-0.151 |
| 83 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.467 |
-0.087 |
| 84 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
-0.458 |
0.139 |
| 85 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.411 |
0.353 |
| 86 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.408 |
0.114 |
| 87 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.408 |
0.293 |
| 88 |
250327_at |
AT5G12050
|
unknown |
-0.400 |
0.173 |
| 89 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.400 |
-0.022 |
| 90 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.399 |
-0.015 |
| 91 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.397 |
-0.038 |
| 92 |
259373_at |
AT1G69160
|
unknown |
-0.391 |
-0.093 |
| 93 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.388 |
0.004 |
| 94 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.387 |
-0.082 |
| 95 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.373 |
-0.070 |
| 96 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.372 |
0.144 |
| 97 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.368 |
0.194 |
| 98 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.366 |
-0.003 |
| 99 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.356 |
0.027 |
| 100 |
257026_at |
AT3G19200
|
unknown |
-0.354 |
0.035 |
| 101 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.353 |
0.176 |
| 102 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.349 |
0.029 |
| 103 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.339 |
-0.051 |
| 104 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
-0.333 |
-0.111 |
| 105 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
-0.332 |
0.156 |
| 106 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.330 |
0.215 |
| 107 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.327 |
0.123 |
| 108 |
260143_at |
AT1G71880
|
ATSUC1, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 1, SUC1, sucrose-proton symporter 1 |
-0.326 |
0.022 |
| 109 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-0.325 |
-0.052 |
| 110 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.323 |
-0.544 |
| 111 |
247474_at |
AT5G62280
|
unknown |
-0.310 |
0.102 |
| 112 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.310 |
0.096 |
| 113 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
-0.305 |
-0.042 |
| 114 |
255788_at |
AT2G33310
|
IAA13, auxin-induced protein 13 |
-0.298 |
0.123 |
| 115 |
245076_at |
AT2G23170
|
GH3.3 |
-0.297 |
0.091 |
| 116 |
248801_at |
AT5G47370
|
HAT2 |
-0.294 |
0.010 |
| 117 |
260264_at |
AT1G68500
|
unknown |
-0.293 |
-0.004 |
| 118 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.292 |
0.088 |
| 119 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.292 |
-0.028 |
| 120 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.292 |
-0.034 |
| 121 |
266831_at |
AT2G22830
|
SQE2, squalene epoxidase 2 |
-0.280 |
0.011 |