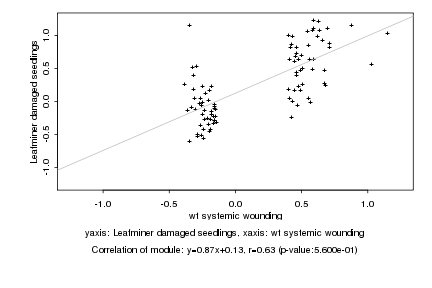
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
wt systemic wounding |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.142 |
1.046 |
| 2 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.023 |
0.576 |
| 3 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.873 |
1.160 |
| 4 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.709 |
0.889 |
| 5 |
262756_at |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
0.704 |
0.830 |
| 6 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.691 |
1.110 |
| 7 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.680 |
0.255 |
| 8 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.672 |
0.283 |
| 9 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.671 |
0.484 |
| 10 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.656 |
0.932 |
| 11 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.629 |
1.092 |
| 12 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.625 |
1.222 |
| 13 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.615 |
0.998 |
| 14 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.588 |
0.649 |
| 15 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.585 |
1.244 |
| 16 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.584 |
1.117 |
| 17 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.582 |
1.085 |
| 18 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.582 |
0.488 |
| 19 |
246911_at |
AT5G25810
|
tny, TINY |
0.560 |
-0.000 |
| 20 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.554 |
0.643 |
| 21 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.550 |
0.865 |
| 22 |
260264_at |
AT1G68500
|
unknown |
0.548 |
0.057 |
| 23 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.538 |
1.077 |
| 24 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.505 |
0.513 |
| 25 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.500 |
0.267 |
| 26 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.499 |
0.699 |
| 27 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.489 |
0.180 |
| 28 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.487 |
0.474 |
| 29 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.475 |
0.235 |
| 30 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.470 |
0.651 |
| 31 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.466 |
-0.047 |
| 32 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.461 |
0.398 |
| 33 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.458 |
0.826 |
| 34 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.456 |
0.734 |
| 35 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.454 |
0.448 |
| 36 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.447 |
0.697 |
| 37 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
0.439 |
0.174 |
| 38 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.439 |
0.610 |
| 39 |
247464_at |
AT5G62070
|
IQD23, IQ-domain 23 |
0.429 |
0.012 |
| 40 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.428 |
0.989 |
| 41 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.423 |
-0.233 |
| 42 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.419 |
0.876 |
| 43 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.410 |
0.824 |
| 44 |
261318_at |
AT1G53035
|
unknown |
0.405 |
0.051 |
| 45 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.403 |
0.640 |
| 46 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.396 |
1.003 |
| 47 |
249411_at |
AT5G40390
|
RS5, raffinose synthase 5, SIP1, seed imbibition 1-like |
0.395 |
0.190 |
| 48 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.387 |
0.265 |
| 49 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.364 |
-0.129 |
| 50 |
263182_at |
AT1G05575
|
unknown |
-0.352 |
1.162 |
| 51 |
250265_at |
AT5G12900
|
unknown |
-0.352 |
-0.600 |
| 52 |
254954_at |
AT4G10910
|
unknown |
-0.335 |
-0.076 |
| 53 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.326 |
0.518 |
| 54 |
255654_at |
AT4G00970
|
CRK41, cysteine-rich RLK (RECEPTOR-like protein kinase) 41 |
-0.319 |
0.396 |
| 55 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.318 |
0.187 |
| 56 |
260206_at |
AT1G70740
|
[Protein kinase superfamily protein] |
-0.311 |
0.053 |
| 57 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.306 |
-0.109 |
| 58 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.300 |
0.544 |
| 59 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.290 |
-0.517 |
| 60 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.289 |
-0.492 |
| 61 |
252501_at |
AT3G46880
|
unknown |
-0.277 |
-0.024 |
| 62 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.266 |
-0.363 |
| 63 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.265 |
0.052 |
| 64 |
250975_at |
AT5G03050
|
unknown |
-0.260 |
-0.046 |
| 65 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.257 |
-0.503 |
| 66 |
265189_at |
AT1G23840
|
unknown |
-0.256 |
0.232 |
| 67 |
257848_at |
AT3G13030
|
[hAT transposon superfamily protein] |
-0.251 |
-0.010 |
| 68 |
255957_at |
AT1G22160
|
unknown |
-0.251 |
-0.196 |
| 69 |
265869_at |
AT2G01760
|
ARR14, response regulator 14, RR14, response regulator 14 |
-0.243 |
-0.412 |
| 70 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.242 |
-0.548 |
| 71 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.237 |
-0.266 |
| 72 |
250720_at |
AT5G06180
|
unknown |
-0.236 |
-0.132 |
| 73 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
-0.233 |
0.136 |
| 74 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.217 |
-0.247 |
| 75 |
258935_at |
AT3G10120
|
unknown |
-0.210 |
0.019 |
| 76 |
251278_at |
AT3G61780
|
emb1703, embryo defective 1703 |
-0.204 |
-0.334 |
| 77 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.201 |
-0.448 |
| 78 |
249221_at |
AT5G42440
|
[Protein kinase superfamily protein] |
-0.200 |
0.173 |
| 79 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.194 |
-0.418 |
| 80 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.189 |
-0.258 |
| 81 |
250823_at |
AT5G05180
|
unknown |
-0.187 |
-0.140 |
| 82 |
249550_at |
AT5G38210
|
[Protein kinase family protein] |
-0.186 |
0.235 |
| 83 |
253980_at |
AT4G26620
|
[Sucrase/ferredoxin-like family protein] |
-0.182 |
-0.183 |
| 84 |
265873_at |
AT2G01630
|
[O-Glycosyl hydrolases family 17 protein] |
-0.169 |
-0.227 |
| 85 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
-0.169 |
-0.327 |
| 86 |
249216_at |
AT5G42240
|
scpl42, serine carboxypeptidase-like 42 |
-0.164 |
-0.105 |
| 87 |
255487_at |
AT4G02610
|
[Aldolase-type TIM barrel family protein] |
-0.161 |
-0.075 |
| 88 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.161 |
-0.044 |
| 89 |
256706_at |
AT3G30300
|
[O-fucosyltransferase family protein] |
-0.159 |
-0.274 |
| 90 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
-0.159 |
-0.276 |
| 91 |
253185_at |
AT4G35240
|
unknown |
-0.158 |
-0.225 |
| 92 |
245074_at |
AT2G23200
|
[Protein kinase superfamily protein] |
-0.155 |
-0.115 |
| 93 |
264361_at |
AT1G03300
|
unknown |
-0.148 |
-0.310 |