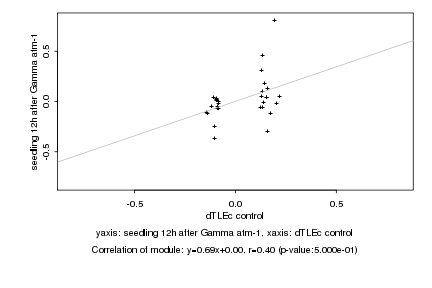
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
dTLEc control |
Log2 signal ratio
seedling 12h after Gamma atm-1 |
| 1 |
255845_at |
AT2G33600
|
AtCRL2, CRL2, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 2 |
0.215 |
0.058 |
| 2 |
266854_at |
AT2G26940
|
C2H2-type zinc finger family protein |
0.203 |
-0.018 |
| 3 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.193 |
0.816 |
| 4 |
247882_at |
AT5G57785
|
unknown |
0.169 |
-0.115 |
| 5 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.156 |
-0.289 |
| 6 |
263919_at |
AT2G36470
|
unknown |
0.156 |
0.130 |
| 7 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
0.153 |
0.047 |
| 8 |
250349_at |
AT5G12000
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.152 |
0.047 |
| 9 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
0.140 |
0.184 |
| 10 |
246504_at |
AT5G16160
|
unknown |
0.135 |
-0.007 |
| 11 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
0.134 |
0.103 |
| 12 |
261318_at |
AT1G53035
|
unknown |
0.132 |
0.466 |
| 13 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.131 |
-0.059 |
| 14 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.127 |
0.313 |
| 15 |
248647_at |
AT5G49190
|
ATSUS2, SSA, SUCROSE SYNTHASE FROM ARABIDOPSIS, SUS2, sucrose synthase 2 |
0.126 |
0.053 |
| 16 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.123 |
-0.057 |
| 17 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.146 |
-0.109 |
| 18 |
265339_at |
AT2G18230
|
AtPPa2, pyrophosphorylase 2, PPa2, pyrophosphorylase 2 |
-0.140 |
-0.116 |
| 19 |
254614_at |
AT4G19191
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.121 |
-0.040 |
| 20 |
246815_at |
AT5G27220
|
[Frigida-like protein] |
-0.110 |
0.042 |
| 21 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
-0.108 |
-0.244 |
| 22 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.107 |
-0.360 |
| 23 |
262650_at |
AT1G14090
|
[pseudogene] |
-0.098 |
0.024 |
| 24 |
262739_at |
AT1G28650
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.097 |
0.040 |
| 25 |
257358_at |
AT2G40990
|
[DHHC-type zinc finger family protein] |
-0.094 |
0.016 |
| 26 |
261095_at |
AT1G62930
|
RPF3, RNA processing factor 3 |
-0.093 |
-0.069 |
| 27 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
-0.090 |
-0.060 |
| 28 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.090 |
-0.043 |
| 29 |
266917_at |
AT2G45830
|
DTA2, downstream target of AGL15 2 |
-0.087 |
-0.069 |
| 30 |
262687_at |
AT1G62670
|
RPF2, rna processing factor 2 |
-0.086 |
0.005 |
| 31 |
265094_at |
AT1G03890
|
[RmlC-like cupins superfamily protein] |
-0.085 |
-0.015 |