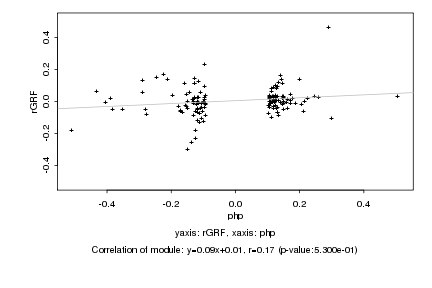
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
php |
Log2 signal ratio
rGRF |
| 1 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
0.504 |
0.034 |
| 2 |
246888_at |
AT5G26270
|
unknown |
0.297 |
-0.103 |
| 3 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.288 |
0.466 |
| 4 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.256 |
0.029 |
| 5 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.246 |
0.037 |
| 6 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
0.224 |
0.022 |
| 7 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
0.214 |
0.004 |
| 8 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.210 |
-0.057 |
| 9 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.203 |
-0.018 |
| 10 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.199 |
0.144 |
| 11 |
262322_at |
AT1G27590
|
unknown |
0.187 |
-0.008 |
| 12 |
251292_at |
AT3G61920
|
unknown |
0.175 |
0.019 |
| 13 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.171 |
0.010 |
| 14 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.171 |
-0.010 |
| 15 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
0.169 |
0.044 |
| 16 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.162 |
0.017 |
| 17 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
0.160 |
-0.042 |
| 18 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.158 |
0.000 |
| 19 |
257527_at |
AT3G01880
|
unknown |
0.149 |
-0.044 |
| 20 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.148 |
0.028 |
| 21 |
252213_at |
AT3G50210
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.147 |
0.032 |
| 22 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.147 |
0.002 |
| 23 |
265005_at |
AT1G61667
|
unknown |
0.147 |
-0.010 |
| 24 |
250028_at |
AT5G18130
|
unknown |
0.146 |
0.115 |
| 25 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.145 |
-0.016 |
| 26 |
248091_at |
AT5G55120
|
galactose-1-phosphate guanylyltransferase (GDP)s;GDP-D-glucose phosphorylases;quercetin 4'-O-glucosyltransferases |
0.145 |
0.022 |
| 27 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
0.144 |
0.034 |
| 28 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.141 |
0.139 |
| 29 |
255286_at |
AT4G04650
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
0.140 |
-0.005 |
| 30 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.140 |
0.164 |
| 31 |
245728_at |
AT1G73340
|
[Cytochrome P450 superfamily protein] |
0.133 |
0.012 |
| 32 |
264882_at |
AT1G61110
|
anac025, NAC domain containing protein 25, NAC025, NAC domain containing protein 25 |
0.133 |
-0.083 |
| 33 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
0.132 |
0.123 |
| 34 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
0.130 |
-0.063 |
| 35 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
0.129 |
-0.064 |
| 36 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.128 |
-0.033 |
| 37 |
246388_at |
AT1G77405
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.128 |
0.034 |
| 38 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
0.128 |
0.095 |
| 39 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.126 |
0.084 |
| 40 |
245864_at |
AT1G58070
|
unknown |
0.126 |
0.011 |
| 41 |
253513_at |
AT4G31760
|
[Peroxidase superfamily protein] |
0.126 |
-0.038 |
| 42 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.124 |
0.031 |
| 43 |
265012_at |
AT1G24470
|
ATKCR2, KCR2, beta-ketoacyl reductase 2 |
0.124 |
0.106 |
| 44 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
0.123 |
0.042 |
| 45 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
0.122 |
0.012 |
| 46 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.122 |
-0.023 |
| 47 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
0.120 |
0.000 |
| 48 |
250483_at |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
0.118 |
0.013 |
| 49 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.118 |
0.092 |
| 50 |
249209_at |
AT5G42620
|
[metalloendopeptidases] |
0.118 |
0.004 |
| 51 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.116 |
0.033 |
| 52 |
250301_at |
AT5G11970
|
unknown |
0.116 |
-0.030 |
| 53 |
257672_at |
AT3G20300
|
unknown |
0.116 |
-0.039 |
| 54 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
0.116 |
0.010 |
| 55 |
256710_at |
AT3G30350
|
GLV3, GOLVEN 3, RGF4, root meristem growth factor 4 |
0.114 |
-0.003 |
| 56 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
0.113 |
0.036 |
| 57 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.111 |
0.086 |
| 58 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
0.110 |
-0.098 |
| 59 |
256935_at |
AT3G22570
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.110 |
0.065 |
| 60 |
260979_at |
AT1G53510
|
ATMPK18, ARABIDOPSIS THALIANA MAP KINASE 18, MPK18, mitogen-activated protein kinase 18 |
0.109 |
0.001 |
| 61 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.109 |
-0.012 |
| 62 |
248549_at |
AT5G50310
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.108 |
-0.007 |
| 63 |
250874_at |
AT5G04010
|
[F-box family protein] |
0.107 |
0.026 |
| 64 |
258561_at |
AT3G05960
|
ATSTP6, SUGAR TRANSPORTER 6, STP6, sugar transporter 6 |
0.106 |
-0.024 |
| 65 |
248116_at |
AT5G55020
|
ATMYB120, MYB120, myb domain protein 120 |
0.105 |
-0.032 |
| 66 |
250231_at |
AT5G13910
|
LEP, LEAFY PETIOLE |
0.105 |
0.001 |
| 67 |
252685_at |
AT3G44410
|
[pseudogene] |
0.104 |
0.028 |
| 68 |
254583_at |
AT4G19480
|
unknown |
0.104 |
0.033 |
| 69 |
247155_at |
AT5G65750
|
[2-oxoglutarate dehydrogenase, E1 component] |
0.104 |
0.039 |
| 70 |
252623_at |
AT3G45320
|
unknown |
0.103 |
0.021 |
| 71 |
263923_at |
AT2G36485
|
[ENTH/VHS family protein] |
0.101 |
-0.071 |
| 72 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.101 |
-0.024 |
| 73 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.513 |
-0.180 |
| 74 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.433 |
0.068 |
| 75 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.405 |
-0.005 |
| 76 |
256928_at |
AT3G22590
|
CDC73, PHP, PLANT HOMOLOGOUS TO PARAFIBROMIN |
-0.392 |
0.021 |
| 77 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.385 |
-0.046 |
| 78 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.353 |
-0.044 |
| 79 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
-0.291 |
0.134 |
| 80 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.290 |
0.057 |
| 81 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.282 |
-0.044 |
| 82 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.280 |
-0.075 |
| 83 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.246 |
0.151 |
| 84 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.225 |
0.174 |
| 85 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.213 |
0.143 |
| 86 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.198 |
0.040 |
| 87 |
247111_at |
AT5G65880
|
unknown |
-0.178 |
-0.028 |
| 88 |
249612_at |
AT5G37290
|
[ARM repeat superfamily protein] |
-0.172 |
-0.062 |
| 89 |
250537_at |
AT5G08565
|
[Transcription initiation Spt4-like protein] |
-0.172 |
-0.055 |
| 90 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
-0.168 |
-0.066 |
| 91 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.159 |
0.115 |
| 92 |
259385_at |
AT1G13470
|
unknown |
-0.157 |
-0.019 |
| 93 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.154 |
-0.026 |
| 94 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.153 |
0.050 |
| 95 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
-0.152 |
0.002 |
| 96 |
256096_at |
AT1G13650
|
unknown |
-0.150 |
-0.294 |
| 97 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
-0.150 |
-0.039 |
| 98 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.146 |
0.061 |
| 99 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.139 |
-0.254 |
| 100 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.134 |
0.018 |
| 101 |
259062_at |
AT3G07440
|
unknown |
-0.133 |
-0.007 |
| 102 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
-0.133 |
0.001 |
| 103 |
254201_at |
AT4G24130
|
unknown |
-0.131 |
-0.086 |
| 104 |
246254_at |
AT4G36450
|
ATMPK14, mitogen-activated protein kinase 14, MPK14, MPK14, mitogen-activated protein kinase 14 |
-0.130 |
0.025 |
| 105 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.129 |
0.148 |
| 106 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
-0.129 |
0.117 |
| 107 |
254423_at |
AT4G21610
|
LOL2, lsd one like 2 |
-0.129 |
0.030 |
| 108 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.129 |
0.117 |
| 109 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.127 |
-0.179 |
| 110 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.127 |
-0.230 |
| 111 |
258867_at |
AT3G03130
|
unknown |
-0.125 |
-0.058 |
| 112 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.122 |
-0.018 |
| 113 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
-0.121 |
-0.118 |
| 114 |
256959_at |
AT3G13440
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.121 |
0.028 |
| 115 |
249362_at |
AT5G40550
|
AtSGF29b, SGF29b, SaGa associated Factor 29 b |
-0.121 |
-0.049 |
| 116 |
265886_at |
AT2G25620
|
AtDBP1, DNA-binding protein phosphatase 1, DBP1, DNA-binding protein phosphatase 1 |
-0.121 |
-0.068 |
| 117 |
260640_at |
AT1G53350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.119 |
0.003 |
| 118 |
245367_at |
AT4G16265
|
NRPB9B, NRPD9B, NRPE9B |
-0.119 |
-0.041 |
| 119 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-0.118 |
-0.002 |
| 120 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.116 |
0.130 |
| 121 |
267205_at |
AT2G30820
|
unknown |
-0.116 |
0.027 |
| 122 |
254039_at |
AT4G25840
|
GPP1, glycerol-3-phosphatase 1 |
-0.116 |
-0.007 |
| 123 |
265907_at |
AT2G25650
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.113 |
-0.072 |
| 124 |
247432_at |
AT5G62500
|
ATEB1, ATEB1B, END BINDING PROTEIN 1B, EB1B, end binding protein 1B |
-0.112 |
-0.072 |
| 125 |
263674_at |
AT2G04790
|
unknown |
-0.112 |
-0.129 |
| 126 |
259210_at |
AT3G09150
|
ATHY2, ARABIDOPSIS ELONGATED HYPOCOTYL 2, GUN3, GENOMES UNCOUPLED 3, HY2, ELONGATED HYPOCOTYL 2 |
-0.111 |
-0.042 |
| 127 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.111 |
0.059 |
| 128 |
261087_at |
AT1G17350
|
[NADH:ubiquinone oxidoreductase intermediate-associated protein 30] |
-0.107 |
-0.012 |
| 129 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.107 |
-0.103 |
| 130 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.106 |
-0.036 |
| 131 |
245426_at |
AT4G17540
|
unknown |
-0.103 |
-0.061 |
| 132 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
-0.102 |
-0.001 |
| 133 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.101 |
-0.121 |
| 134 |
256512_at |
AT1G33265
|
[Transmembrane proteins 14C] |
-0.100 |
0.001 |
| 135 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.099 |
0.017 |
| 136 |
257762_at |
AT3G23100
|
XRCC4, homolog of X-ray repair cross complementing 4 |
-0.099 |
-0.032 |
| 137 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.099 |
0.235 |
| 138 |
254094_at |
AT4G25120
|
ATSRS2, ARABIDOPSIS THALIANA SUPPRESSOR OF RAD SIX-SCREEN MUTANT 2, SRS2, SUPPRESSOR OF RAD SIX-SCREEN MUTANT 2 |
-0.098 |
0.096 |
| 139 |
265852_at |
AT2G42350
|
[RING/U-box superfamily protein] |
-0.098 |
0.026 |
| 140 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.096 |
-0.012 |
| 141 |
251215_at |
AT3G62510
|
[protein disulfide isomerase-related] |
-0.096 |
-0.015 |
| 142 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.095 |
-0.086 |
| 143 |
266064_at |
AT2G18780
|
[F-box and associated interaction domains-containing protein] |
-0.095 |
0.038 |